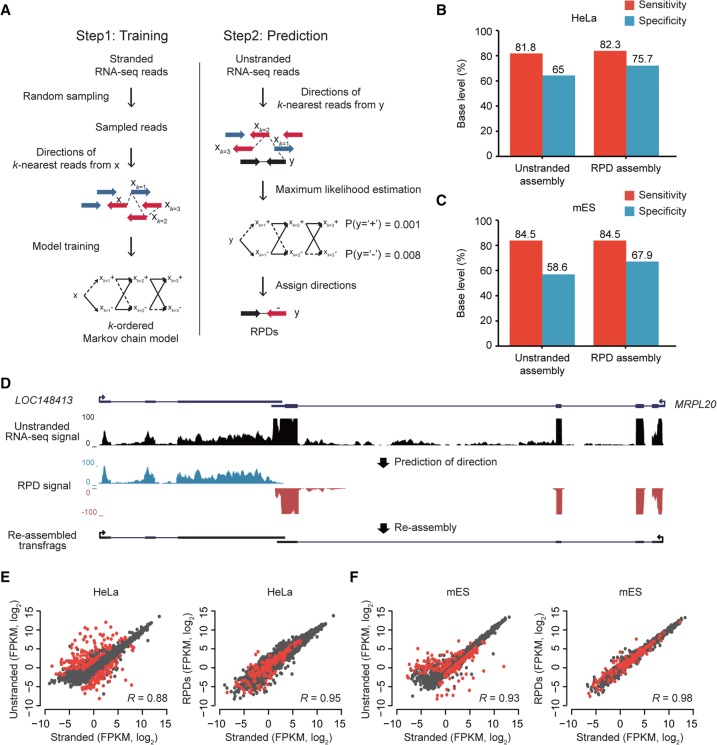
Figure 2.
Prediction of read directions using MLE. (A) Overview of kMC training and MLE of read direction. (Left) S base reads randomly sampled from stranded RNA-seq reads and their matched step-wise k-nearest reads (xk=1, xk = 2, xk=3,…) were used for training kMC. Blue arrows are reads in the forward (+) direction, and red arrows are reads in the reverse (−) direction. (Right) Prediction of read direction using MLE. Step-wise k-nearest stranded reads (xk=1, xk = 2, xk=3,…) from a query unstranded read (black arrow) were extracted and used to calculate two likelihoods at (+) and (−). A direction with the maximum likelihood is finally assigned to the query read. (B,C) Accuracies of transcriptomes assembled with RPDs (k = 3) and unstranded reads in HeLa (B) and mES cells (C). (D) An example of resulting transfrags reassembled with RPDs. LOC148413 and MRPL20 are convergently overlapped at a locus where unstranded RNA-seq signals (black) are not separated, but blue and red RPD signals are clearly separated in the forward and reverse directions, respectively. (E,F). Comparisons of gene expression values (FPKM, log2) estimated by stranded (x-axis) and unstranded reads (y-axis, left) or RPDs (y-axis, right) in HeLa (E) and mES cells (F). The correlation coefficients were calculated with Pearson's correlation between the x- and y-axis values. The red dots indicate genes with antisense-overlapped genes.