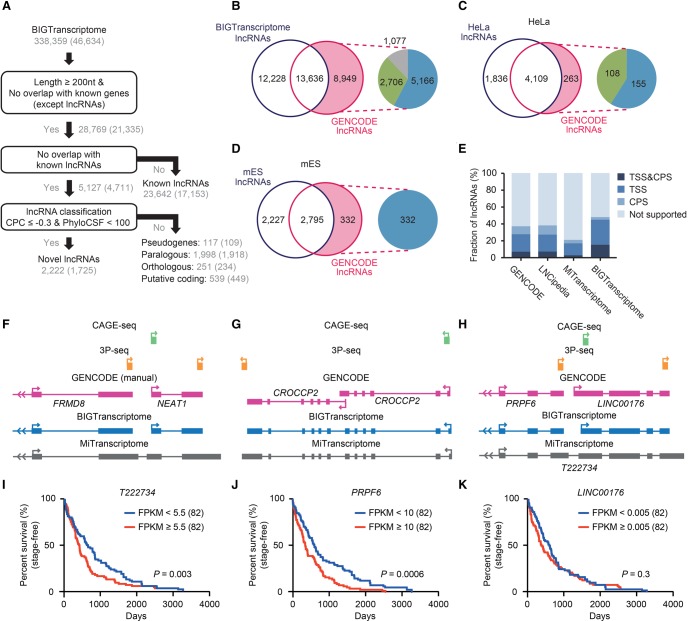
Figure 7.
BIGTranscriptome includes known and novel noncoding genes. (A) A schematic flow for annotating novel and known noncoding genes in BIGTranscriptome. (B) The Venn diagrams display the fraction of BIGTranscriptome lncRNAs that are published GENCODE lncRNAs. The inset indicates that GENCODE lncRNAs (8949) not detected in BIGTranscriptome were classified as overlapping with known genes (blue), overlapping with falsely fused genes (green), or truly missed in our catalog (gray). (C,D) Transcriptomes of HeLa (C) and mES cells (D) were compared to GENCODE lncRNAs, expressed over 1 FPKM in the matched cell types. The insets indicate that HeLa- and mES-expressed lncRNAs not detected in our lncRNA set were filtered by either overlap with known genes (blue) or misannotation (green). (E) The fractions of the indicated lncRNA sets with both TSS and CPS, either site, or neither site are shown in bar graphs. (F–H) Examples of misannotated gene models in public databases (MiTranscriptome and GENCODE). (F) The gene for a well-studied lncRNA, NEAT1, has been combined with a protein-coding gene, FRMD8, leading to misannotation as a protein-coding gene. (G) CROCCP2 is annotated in GENCODE (automatic) as having two independent isoforms whereas it is annotated as a single transcript in BIGTranscriptome and MiTranscriptome. (H) Gene models of BIGTranscriptome and MiTranscriptome, and CAGE-seq and 3P-seq data, at a locus. A fused single form, T222734, was annotated in MiTranscriptome whereas two independent genes, PRPF6 and LINC00176, were annotated in BIGTranscriptome. (I–K) Survival analyses for TCGA liver cancer samples based on the resulting gene models. One hundred sixty-four patient samples including termination events were divided into two groups, the top 50% (red) and bottom 50% (blue), by the median FPKM values of T222834 (I), PRPF6 (J), and LINC00176 (K).