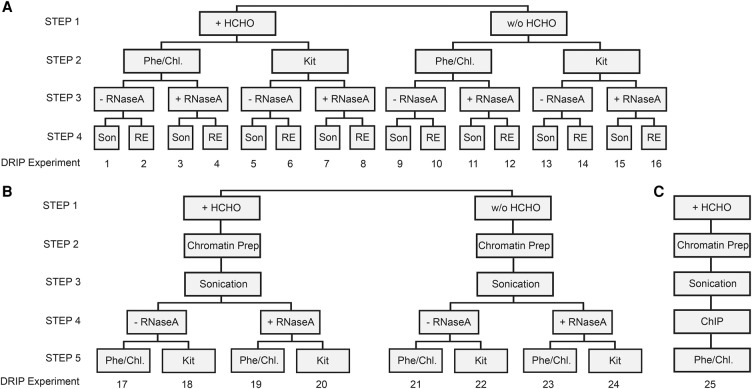
Figure 1.
Experimental design: constructing DRIP schemes. (A) Experiments 1–16 explore the effect of formaldehyde-fixation (Step 1), nucleic acid isolation (Step 2), removal of free RNA (Step 3), and nucleic acid fragmentation (Step 4) on the outcome of RNA-DNA hybrid detection. Each experiment was performed at two parallel cell lysis temperatures (65°C and 37°C), respectively. The temperature variable is not depicted in the cartoon, but it is referred in the main text. (B) Experiments 17–24 test the impact of acoustic sharing performed on a chromatin prep rather than on naked nucleic acid, similarly to the ChIP protocol. Each experiment was performed at 65°C cell lysis temperature. (C) Workflow of a ChIP experiment (shown only for comparison with the DRIP pipeline). (HCHO) Formaldehyde fixation, (Phe/Chl) phenol-chloroform extraction, (Kit) silica membrane-based nucleic acid purification, (RNase A) Ribonuclease A digestion performed at high (300 mM) NaCl concentration, (Son) sonication, (RE) restriction enzyme cocktail digestion (HindIII, EcoRI, BsrGI, XbaI, and SspI). As a negative control, RNase H digestion was applied in all DRIP experiments (not indicated in the cartoon).