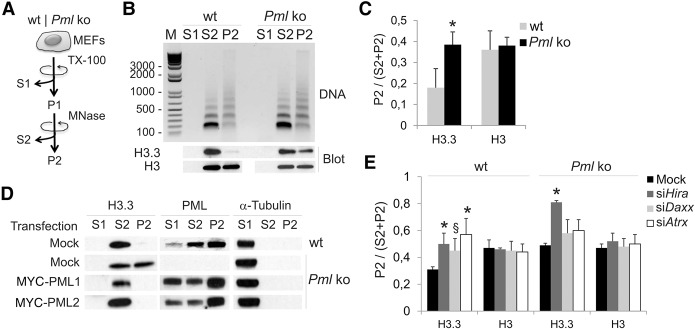
Figure 1.
Loss of PML partitions histone H3.3 into a compact micrococcal nuclease (MNase)–resistant chromatin compartment. (A) MEF fractionation into a cytosolic fraction (S1), a Triton X-100 (0.5%)-insoluble/MNase (0.5 U/µg DNA)-soluble fraction (S2), and a Triton X-100/MNase-insoluble fraction (P2). (B) Nucleosome laddering (DNA; top) and Western blot (bottom) of endogenous H3.3 and total H3 in each fraction, from wt and Pml ko MEFs. (C) Proportions of H3.3 and total H3 in P2; (*) P < 0.05 relative to wt; paired t-tests. (D) Distribution of H3.3 and PML in S1, S2, and P2 from mock-transfected wt and Pml ko MEFs and from ko MEFs expressing MYC-PML1 or MYC-PML2 (full-length PML); alpha-tubulin was used as loading control for S1. (E) Proportions of H3.3 and total H3 in P2 after knockdown of Hira, Daxx, or Atrx; mean ± SD, three experiments; (*) P = 0.03 (siHira wt); 0.04 (siAtrx wt); 1.4 × 10−5 (siHira ko); (§) P = 0.06 (siDaxx, wt); paired t-tests relative to mock control.