Abstract
MicroRNAs (miRNAs) are a group of small RNAs that control gene expression in all aspects of eukaryotic life, primarily through RNA silencing mechanisms. The purpose of the present review is to introduce key miRNAs involved in periodontal homeostasis, summarize the mechanisms by which they affect downstream genes and tissues, and provide an introduction into the therapeutic potential of periodontal miRNAs. In general, miRNAs function synergistically to fine-tune the regulation of biological processes and to remove expression noise rather than by causing drastic changes in expression levels. In the periodontium, miRNAs play key roles in development and periodontal homeostasis and during the loss of periodontal tissue integrity as a result of periodontal disease. As part of the anabolic phase of periodontal homeostasis and periodontal development, miRNAs direct periodontal fibroblasts toward alveolar bone lineage differentiation and new bone formation through WNT, bone morphogenetic protein, and Notch signaling pathways. miRNAs contribute equally to the catabolic aspect of periodontal homeostasis as they affect osteoclastogenesis and osteoclast function, either by directly promoting osteoclast activity or by inhibiting osteoclast signaling intermediaries or through negative feedback loops. Their small size and ability to target multiple regulatory networks of related sets of genes have predisposed miRNAs to become ideal candidates for drug delivery and tissue regeneration. To address the immense therapeutic potential of miRNAs and their antagomirs, an ever growing number of delivery approaches toward clinical applications have been developed, including nanoparticle carriers and secondary structure interference inhibitor systems. However, only a fraction of the miRNAs involved in periodontal health and disease are known today. It is anticipated that continued research will lead to a more comprehensive understanding of the periodontal miRNA world, and a systematic effort toward harnessing the enormous therapeutic potential of these small molecules will greatly benefit the future of periodontal patient care.
Keywords: small RNA, periodontium, osteoblast, osteoclast, alveolar bone, nanoparticle
Periodontal Tissue Homeostasis
The periodontal region is a highly dynamic microenvironment that undergoes continuous remodeling due to frequent tissue turnover, high levels of mechanical stress, and inflammatory conditions in tissues affected by periodontal disease. The remarkably high rate of collagen turnover and extracellular matrix remodeling in periodontal tissues has been well documented (Kameyama 1973; Rippin 1976, 1978; Sodek and Ferrier 1988; Bartold 1995). In periodontally healthy individuals, the rates of matrix degradation and bone resorption are in balance with the rates of new matrix formation and new bone deposition, a state called homeostasis. This physiological equilibrium between bone resorption and new bone formation that results in the maintenance of tissue morphology while allowing for continuous turnover is also termed coupling (Mohri et al. 1991; Graves et al. 2011).
The key cells responsible for periodontal tissue homeostasis are the periodontal progenitors (PDLSCs), a group of tissue-specific stem cells that are capable of forming new periodontal ligament (PDL) (Dangaria et al. 2011a; Dangaria, Ito, Yin, et al. 2011). Periodontal tissues originate from neural crest–derived intermediate progenitors of the dental follicle that give rise to PDL fibroblasts, alveolar bone osteoblasts, and cementoblasts (Diekwisch 2002; Luan et al. 2009; Dangaria et al. 2011b). These periodontal progenitors not only maintain the nonmineralized PDL but also the integrity of the mineralized alveolar socket, which anchors the teeth within jaws (Dangaria et al. 2009, 2011b; Jung et al. 2011). The common mineralized tissue lineage origin of mammalian periodontal progenitors has been demonstrated by marker studies for the early mineralization marker RunX2 (Luan et al. 2006, 2009), suggesting that subsequent periodontal tissue differentiation involves finely tuned spatial control of mineralization. The segregation of the periodontal attachment apparatus into mineralized and nonmineralized components is a unique attribute of mammals and rarely occurs in other animals (Diekwisch 2016a), indicating that mineralized state homeostasis in the periodontal region has evolved over millions of years throughout the course of vertebrate evolution (McIntosh et al. 2002). In healthy mammalian periodontia, alveolar bone osteoblasts/osteocytes continuously deposit new mineralized alveolar bone tissue and collagenous extracellular matrix to offset the loss of bone and matrix as part of the physiological remodeling process.
In patients with periodontal disease, the balance between anabolic and catabolic processes is disturbed, causing increased resorptive activity, decreased new bone formation, and an incomplete deposition of new matrix into recent resorption lacunae, a process called “uncoupling” (Redlich and Smolen 2012). Periodontal disease originates from a microbial challenge to the gingival tissues that is based on a dysbiotic microflora of periodontal pathogens (Löe et al. 1965; Theilade et al. 1966; Page and Schroeder 1976). These periodontal pathogens reside within the supra- and subgingival plaque and cause an inflammatory response in gingival tissues through the activation of prostaglandins, cytokines, and chemokines (Darveau 2010; Graves et al. 2011). When periodontal disease progresses, the initial inflammatory response is followed by a breakdown of periodontal connective tissue extracellular matrices, including alveolar bone, in tandem with an exacerbation of the inflammatory reaction and corresponding host response (Graves 2008; Bartold and Van Dyke 2013). While the precise trigger mechanisms of the transition from gingivitis to periodontitis remain to be defined, recent studies have demonstrated that the periodontal host response plays a predominant role during the pathogenesis of periodontitis (Graves et al. 2011; Marsh and Devine 2011; Bartold and Van Dyke 2013). Moreover, during periodontitis progression, the virulence of bacterial pathogens and the severity of the inflammatory response potentiate each other, causing an escalation of periodontal tissue destruction and eventually tooth loss (Hajishengallis 2014; Lamont and Hajishengallis 2015).
All aspects of this process, from the initial inflammatory reaction in the gingiva to the impairment of bone and matrix synthesis and the escalation of matrix and bone destruction, are governed by a group of small RNAs called microRNAs (miRNAs) that only recently have emerged as the most important regulators of bone formation, resorption, remodeling, repair, and disease (Zhao et al. 2016).
MicroRNA Biogenesis and Function
MicroRNAs are 21- to 23-nucleotide noncoding RNA molecules that are involved in almost all aspects of eukaryotic life. In recent years, miRNAs have emerged as regulators of a multitude of discrete biological processes involved in the cross-coordination and functional integration of complex physiological events (Millar et al. 2015). Moreover, it has been demonstrated that miRNAs have multiple roles in all aspects of periodontal tissue homeostasis, including periodontal stem cell differentiation, osteoblast and osteoclast function, and response to mechanical stress (Lian et al. 2012; Irwandi and Vacharaksa 2016).
The process by which miRNAs are formed is called miRNA biogenesis. During this process, miRNAs are first transcribed as primary transcripts (pri-miRNA) with a 5′-cap and a 3′-poly-A tail. The maturation of these primary transcripts involves a 2-step process that begins with pri-miRNA processing by the Drosha ribonuclease into short, 70-nucleotide RNA molecules with stem-loop structures. As a next step, pre-miRNAs are transported from the nucleus to the cytoplasm by the Ran transport receptor Exportin-5. Once in the cytoplasm, pre-miRNAs are further cleaved by an endoribonuclease called Dicer to form a short, double-stranded miRNA:miRNA* duplex. Finally, the miRNA:miRNA* duplex is unwound into mature miRNA and miRNA* single strands by a helicase. The mature miRNAs are then incorporated into the RNA-induced silencing complex (RISC), which initiates RNAi-based gene silencing. A number of recent review articles provide further details related to miRNA biogenesis (Bartel 2004; Carthew and Sontheimer 2009; Cech and Steitz 2014; Ha and Kim 2014).
Most miRNAs affect gene expression via gene silencing mechanisms, including mRNA cleavage and translation repression through the RISC (Hofacker 2007; MacFarlane and Murphy 2010). The type of silencing mechanism employed by an individual miRNA appears to depend on the level of complementarity between the miRNA and mRNA target (Bartel 2004). However, these 2 mechanisms are distinguished by the reversibility of their effect on messenger RNA (mRNA): mRNA decay is an irreversible process, while translation inhibition is reversible because stable mRNA can be translated following elimination of translation repression (Brengues et al. 2005; Maroney et al. 2006; Valencia-Sanchez et al. 2006). While the majority of miRNAs function via silencing mechanisms (Bartel 2004), a number of miRNAs are also involved in the upregulation of gene expression (Orang et al. 2014). Moreover, miRNAs can both regulate and be regulated by target interactions (Pasquinelli 2012), suggesting that miRNAs play multiple roles in complex physiological interactions and disease processes. In the present review, we are summarizing key aspects of miRNA function in periodontal tissue homeostasis and disease progression that are known so far.
miRNA Function in Periodontal Mineralized Tissue Homeostasis
miRNA Regulation of Mineralized Tissue Lineage Commitment
The decision of periodontal lineages to differentiate into alveolar bone osteoblasts/osteocytes, periodontal fibroblasts, or cementoblasts is essential for the life of each individual tooth (Dangaria et al. 2011b). The precise timing and spatial coordination of the mineralization potential that leads to such decision is controlled by RunX2, bone morphogenetic protein (BMP), Notch, and other signaling pathways; and an ever increasing number of miRNAs are involved in the coordination of this process. Here we have conducted a miRNA profiling study that lists individual miRNAs altered as a result of osteogenic induction of periodontal progenitors (Table 1), and in this subsection, we are discussing the potential role of these miRNAs in the context of their known role in osteogenic differentiation.
Table 1.
MicroRNA Changes in Periodontal Progenitors following Osteogenic Induction.
| miRNA | Shift of Expression | miRNA | Shift of Expression |
|---|---|---|---|
| miR-15b | Up | miR-15a | Down |
| miR-17 | Up | miR-34 a,c | Down |
| miR-21 | Up | miR-100 | Down |
| miR-27 | Up | miR-138 | Down |
| miR-29b,c | Up | miR-195 | Down |
| miR-31 | Up | ||
| miR-125a,b | Up | ||
| miR-146a | Up | ||
| miR-199a | Up |
PDLSCs were cultured in control or osteogenic induction conditions for 12 d, and microRNA (miRNA) expression was assessed using a miRNA array.
Our profiling study demonstrated that the expression of miR-31, miR34a, and miR-34c was downregulated during periodontal progenitor differentiation induced by osteogenic conditions. Previous studies established that miR-34c and miR-218 directly target RunX2 (Zhang et al. 2011; Gay et al. 2014), while miR-31 is one of the miRNAs negatively regulated by RunX2 and forms a RunX2/miR-31/Satb2 regulatory loop (Deng et al. 2013). The Special AT-rich sequence-binding protein 2 (SATB2) is a miR-31 target and a nuclear matrix protein critical for osteoblast differentiation (Xie et al. 2014). The 2 miRNAs, miR-31 and miR-34c, affect osteoblastogenesis through the RunX2 pathway. In contrast, miR-15b, which was upregulated in our PDL cell mineralization assay, promotes osteoblast differentiation by indirectly protecting Runx2 protein from Smurf1-mediated degradation (Vimalraj et al. 2014). Smurf1 is a direct target of miR-15b and a ubiquitin protein ligase able to degrade RunX2 by a proteasomal mechanism (Vimalraj et al. 2014). Together, these findings indicate that miRNAs may be either up- or downregulated to affect RunX2-mediated osteogenesis via different and seemingly unrelated mechanisms.
Osteoblast differentiation occurs as the result of the concerted action of multiple signaling pathways, including BMP, Wnt, and Notch pathways. Among these, the BMP signaling pathway plays a fundamental role in osteoblast differentiation. Previous studies demonstrated that BMP signaling is both negatively and positively regulated by miRNAs. In our profiling study, the expression of miR-100 was decreased during osteogenic differentiation of periodontal progenitors (Table 1). Previous studies have revealed that downregulation of miR-100 enhanced osteogenic differentiation of human adipose–derived mesenchymal stem cells (MSCs) and that BMPR2 was a direct target of miR-100 (Zeng et al. 2012). Overexpression of miR-100 reduced BMPR2 gene expression, resulting in an inhibition of RunX2 and bone-related protein expression (Zeng et al. 2012) and suggesting that miR-100 is a negative regulator of osteogenic differentiation. Thus, the decreased expression of miR-100 in our study appears to facilitate osteogenesis because miR-100 acts as a negative regulator of mineralization. The miR-497∼195 cluster is another example of a miRNA-related mechanism involved in the negative regulation of BMP signaling. In our study, miR-195 was downregulated upon osteogenic induction of PDL progenitors (Table 1), and previous studies had demonstrated that miR-195 downregulated the expression of Tgfbr3, Smad5, Mapk3, and Smurf1 and acted as an intracellular antagonist of BMP signaling in bone cells (Grünhagen et al. 2015). This effect may be mediated by chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII), a direct target of miR-195 (Jeong et al. 2014). Thus, the negative regulation of miR-195 in mineralization-induced periodontal progenitors is another example by which an inhibitory miRNA is downregulated to facilitate mineralization induction. miRNAs also facilitate osteogenesis by downregulating inhibitors of mineralization. One example of the repression of inhibitor mode of action is miR-21, which was upregulated in our periodontal progenitor mineralization induction study (Table 1). Based on previous studies, miR-21 was significantly upregulated during BMP9-induced osteogenesis and promoted the osteogenic differentiation of the murine multilineage cells (MMCs) by suppressing Smad7, a negative regulator of MMC osteogenic differentiation (Song et al. 2015). Underscoring the complexity of miRNA function, miR-17 appears to enhance osteogenesis through several mechanisms. By directly targeting Smad7, miR-17 promoted β-catenin activity (Jia et al. 2014), while miR-17 also suppressed the expression of Smurf1 (Liu et al. 2011), a downstream effector of BMP signaling, indicating that miR-17 negatively regulates BMP signaling through various checkpoints. In our study, miR-17 level was upregulated after osteogenesis induction (Table 1).
Wnt signaling has an important function in periodontal development and homeostasis, including the control of PDL width (Lim et al. 2014, 2015; Yin et al. 2015; Diekwisch 2016a; Tamura and Nemoto 2016). Several miRNAs are known to enhance Wnt signaling by targeting Wnt inhibitors and promoting osteoblastogenesis (Li et al. 2009; Wang and Xu 2010). In our periodontal progenitor osteogenic lineage induction study, miR-27, miR-29, and miR-199a were upregulated during osteogenic differentiation of periodontal progenitors (Fig. 1). These miRNAs are known to control osteogenic lineage commitment of various mesenchymal stem cells through the formation of positive feedback loops and Wnt/β-catenin signaling activation. Specifically, miR-27 promotes odontoblast/osteoblast differentiation from odontoblastic progenitor cells by targeting the Wnt-negative regulator adenomatous polyposis coli (Park et al. 2014); miR-29 facilitates osteoblast differentiation from mesenchymal precursor cell line hFOB1.19 by targeting key Wnt signaling antagonists, Dkk1, Kremen2, and sFRP2 (Kapinas et al. 2010); and miR-199 positively regulates osteogenic differentiation of bone marrow stromal cells through suppressing GSK-3β/β-catenin signaling (Zhao et al. 2016). These 3 miRNAs promote osteoblastogenesis by targeting WNT inhibitors, and they are likely to affect mineralized tissue lineage commitment in periodontal progenitors through similar mechanisms.
Figure 1.
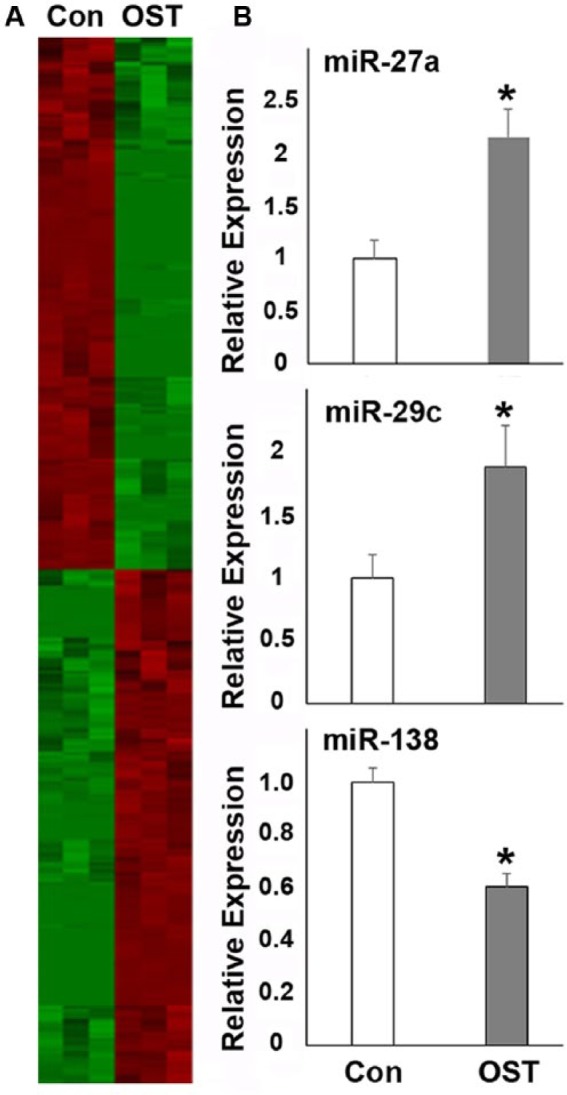
Microarray analysis of microRNA (miRNA) expression in control and osteogenesis-induced PDLSCs. For this study, PDLSCs were cultured under osteogenic induction conditions for 12 d and compared with controls. (A) Heat map of miRNA expression profiling. miRNAs with a significant level of up- or downregulation (P < 0.01) were identified using Student’s t test. Individual up- and downregulated genes are listed in Table 1. (B) quantitative Reverse Transcriptase (qRT)polymerase chain reaction verification of miR-27, miR-29, and miR-138 expression in control and osteogenesis-induced PDLSCs. *P < 0.05.
Notch signaling is a third pathway that is important for MSC differentiation into osteoblasts (Lin and Hankenson 2011). miR-34 family members are inhibitors of Notch signaling and suppress osteoblast differentiation. miR-34a and miR-34c target Notch ligand JAG1 and/or Notch receptors Notch1 and Notch2 during osteoblast differentiation (Bae et al. 2012; Chen, Holmstrøm, et al. 2014). Osteoblast-specific gain of miR-34 function in mice inhibited bone formation. Conversely, miR-34a deficiency increased bone formation by affecting proliferation and mineralization of osteoblasts. These results demonstrate that the miR-34 family plays a critical role in osteoblastogenesis through regulating Notch signaling during bone homeostasis (Bae et al. 2012; Chen, Holmstrøm, et al. 2014).
miRNA Regulation of Osteogenesis under Inflammatory Conditions
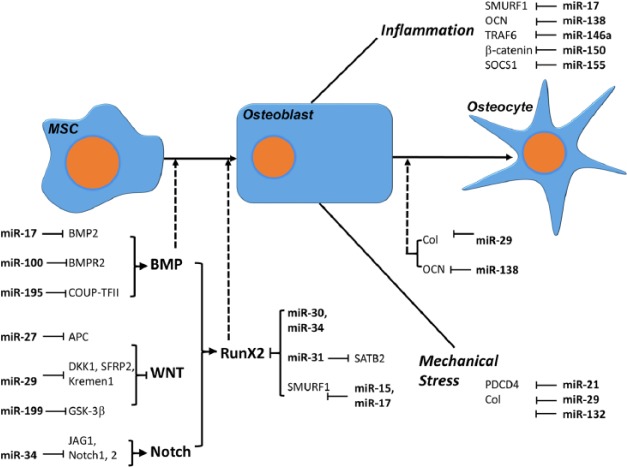
Inflammatory conditions as they occur in periodontal disease often result in alveolar bone loss and disruption of connective tissue homeostasis (Darveau 2010; Bartold and van Dyke 2013; Hienz et al. 2015; Zhou et al. 2016). As mentioned earlier, the etiology of advanced periodontal disease is greatly affected by the host immune response, and it has been demonstrated that osteoblast lineage cells contribute greatly to periodontal bone loss by activating nuclear factor (NF)–κB (Pacios et al. 2015). In this subsection, we have focused on the effect of miRNAs as regulators of periodontal stem cells as they affect homeostasis under inflammatory conditions (Fig. 2).
Figure 2.
MicroRNA (miRNA)–mediated regulation of osteoblastogenesis from mesenchymal stem cells (MSCs). This sketch illustrates the role of miRNAs during the differentiation of mesenchymal stem cells into osteoblasts and osteocytes under physiological and inflammatory conditions or when exposed to mechanical stress. Note that by affecting different types of targeting genes, miRNAs either promote or inhibit osteoblastogenesis.
Our recent study related to the role of miR-138a directly supports the concept of osteoblastic lineage cells as key contributors to periodontal bone loss in inflamed periodontal tissues (Zhou et al. 2016). In this study, we demonstrated that inflammatory conditions significantly increased miR-138 expression, and this increase in turn inhibited key mineralization genes such as osteocalcin (OC), RunX2, and collagen I. Moreover, knockdown of miR-138 or addition of OC protein partially rescued alkaline phosphatase activity in PDL cells subjected to lipopolysaccharide treatment, confirming the essential role of OC in the mineralization of periodontal progenitors (Zhou et al. 2016). These data establish the miR-138 inhibitor as a potential therapeutic agent for the prevention of the bone loss associated with advanced periodontal disease (Zhou et al. 2016).
The effect of miR-17 on osteogenic differentiation in periodontal progenitors from periodontitis-affected patients is another line of evidence that underscores the inhibitory effect of inflammatory environments on periodontal stem cells and osteoblast activity (Liu et al. 2011). This study demonstrates that inflammation inhibits miR-17 expression and promotes Smurf1, a direct target of miR-17 in periodontal progenitors. In a coherent feed-forward loop consisting of inflammatory cytokines, miR-17 and Smurf1, proinflammatory cytokines cause miR-17 downregulation and Smurf1 activation, as well as increased degradation of Smurf1-mediated osteoblast-specific factors (Liu et al. 2011).
Another miRNA that plays a significant role in the differentiation of periodontal ligament cells under inflammatory conditions is miR-146a, which is upregulated during osteogenic differentiation of PDL cells (Hung et al. 2010). In this study, upregulation of miR-146a resulted in attenuation of NF-κB activity and increased osteogenic differentiation marker gene expression profiles, while manipulation of NF-κB activity blocked the function of miR-146a in osteogenesis, indicating that miR-146a promotes the differentiation in PDL cells through the downregulation of NF-κB signaling (Hung et al. 2010).
The miRNAs miR-155 and miR-150 provide further evidence for the role of miRNAs as they contribute to the inhibitory effect of inflammatory conditions on osteoblast differentiation from mesenchymal stem cells and preosteoblasts in vitro. In these studies, proinflammatory conditions were mimicked using tumor necrosis factor (TNF)–α as a proinflammatory cytokine. In a first study, miR-155 modulated TNF-α–inhibited osteogenic differentiation by targeting SOCS1 (Wu et al. 2012), and in a second study, miR-150 modulated TNF-α–inhibited osteogenic differentiation by suppressing β-catenin (Wang et al. 2016). In both cases, the progression of osteogenic differentiation under inflammatory conditions was impeded, either through SAPK/JNK (miR-155; Wu et al. 2012) or through Wnt–β-catenin signaling (miR150; Wang et al. 2016).
miRNA Regulation of Osteoclast Differentiation and Function
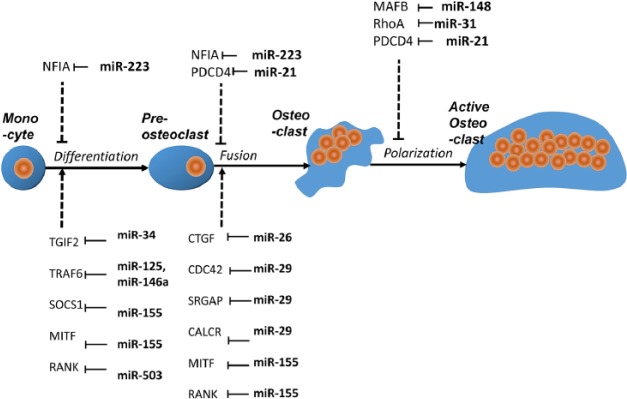
The loss of periodontal homeostasis in inflamed periodontal tissues not only affects the ability of periodontal progenitors and osteoblasts to form new bone but also directly results in bone resorption as a result of osteoclast activation (Hienz et al. 2015). The signaling cascades that lead to osteoclast activation are triggered by lipopolysaccharides from bacterial cells walls and their effect on proinflammatory cytokines such as interleukin (IL)–1β, IL-6, and TNF-α, which in turn stimulate the RANK-ligand (RANKL) receptor (RANK) on the osteoclast cell surface and its nuclear target NF-κB, as well as prompt the differentiation of osteoclast precursors into multinucleated osteoclasts (Wiebe et al. 1996; Boyce 2013; Cekici et al. 2014; Lee et al. 2014; Boyce et al. 2015). Once differentiated, osteoclasts tightly adhere to the bone and secrete a highly acidic solution through their ruffled border into the resorption pit that mobilizes the mineral phase and facilitates organic bone matrix degradation via cathepsin K (Teitelbaum 2000; Karsenty and Wagner 2002).
The importance of miRNAs for osteoclastic activity and function was demonstrated in a mouse model, in which the enzyme necessary for mature miRNA formation following pri-miRNA cleavage (Dicer) was knocked out in osteoclasts by crossing Dicer flox mice with cathepsin K–Cre knock-in mice (Mizoguchi et al. 2010). Knocking out Dicer resulted in decreased osteoclast numbers, a reduction in NFATc1 and TRAP gene expression, and increased bone mass, demonstrating that the cleavage of miRNAs by Dicer is necessary for physiological osteoclast function and bone resorption (Mizoguchi et al. 2010).
Periodontal miRNAs affect the inflammatory activation of osteoclastogenesis and the alveolar bone resorption by periodontal osteoclasts on all levels. Table 2 lists some of the miRNAs expressed in periodontal tissues that are affected by periodontal disease. These miRNAs affect the catabolic aspect of periodontal bone homeostasis either through the inhibition of osteoclast activity and function or by facilitating negative feedback loops or by promoting osteoclast function (Figs. 3, 4).
Table 2.
MicroRNA (miRNA) Changes in Periodontitis Tissues versus Control.
| miRNA | Species | Cell/Tissue | Expression | Reference |
|---|---|---|---|---|
| miR-15a | Human | Gingival tissue | Up |
Perri et al. (2012)
|
| miR-17 | Human | Gingival tissue | Up | Lee et al. (2011); Xie et al. (2011) |
| miR-21 | Rat | Apical tissue | Up | Gao and Zheng (2013) |
| Human | Gingival tissue | Up | Lee et al. (2011) | |
| miR-26 | Human | Gingival tissue | Up | Lee et al. (2011) |
| miR-29a,b,c | Human | Gingival tissue | Up | Lee et al. (2011); Xie et al. (2011); Stoecklin-Wasmer et al. (2012) |
| miR-31 | Human | Gingival tissue | Up | Stoecklin-Wasmer et al. (2012) |
| miR-34a,c | Rat | Apical tissue | Up | Gao and Zheng (2013) |
| Human | Gingival tissue | Up | Lee et al. (2011) | |
| miR-125a | Rat | Apical tissue | Up | Gao and Zheng (2013) |
| Human | Gingival tissue | Up | Lee et al. (2011); Stoecklin-Wasmer et al. (2012) | |
| miR-125b | Human | Gingival tissue | Up | Lee et al. (2011); Xie et al. (2011) |
| miR-132 | Human | Gingival tissue | Up | Kalea et al. (2015) |
| miR-146a | Rat | Apical tissue | Up | Gao and Zheng (2013) |
| Human | Gingival tissue | Up | Xie et al. (2011) | |
| miR-148 | Human | Gingival tissue | Up | Stoecklin-Wasmer et al. (2012) |
| miR-150 | Human | Gingival tissue | Up | Ogata et al. (2014) |
| miR-155 | Human | Gingival tissue | Up | Stoecklin-Wasmer et al. (2012) |
| miR-195 | Human | Gingival tissue | Up | Lee et al. (2011); Xie et al. (2011) |
| miR-223 | Human | Gingival tissue | Up | Ogata et al. (2014); Stoecklin-Wasmer et al. (2012) |
| miR-503 | Rat | Apical tissue | Up | Gao and Zheng (2013) |
| miR-15b | Rat | Apical tissue | Down | Gao and Zheng (2013) |
| miR-17 | Rat | Apical tissue | Down | Gao and Zheng (2013) |
| miR-100 | Human | Gingival tissue | Down | Ogata et al. (2014) |
| miR-132 | Rat | Apical tissue | Down | Gao and Zheng (2013) |
| miR-199a | Human | Gingival tissue | Down | Ogata et al. (2014) |
These data are based on a comparison between gingival tissues of periodontitis patients compared with healthy subjects or on a comparison of apical tissues from a rat apical periodontitis model versus control animals.
Figure 3.
MicroRNA (miRNA)–mediated regulation of osteoclastogenesis. Here we illustrate the effects of miRNAs during the 4 stages of osteoclastogenesis from monocytes to preosteoclasts and then from mature osteoclasts to activated osteoclasts. At each point of transition, microRNAs either promote or inhibit osteoclastogenesis through their targeting genes.
Figure 4.
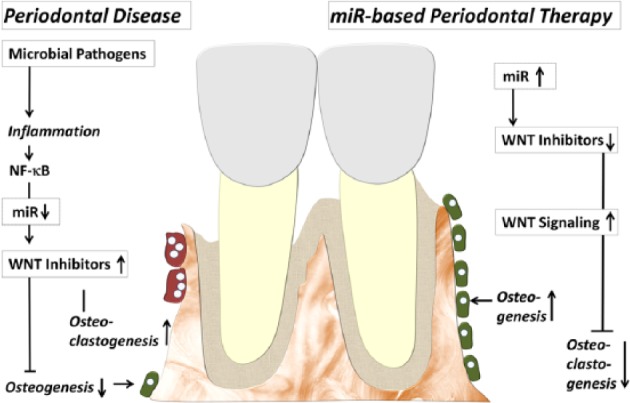
Putative model illustrating the role of microRNAs (miRNAs) and WNT signaling and alveolar bone homeostasis. In this model, inflammatory conditions downregulate miRNA subsets, which leads to an upregulation of some WNT inhibitors, a reduction in osteogenesis, and an increase in bone resorption. Therapy with selected miRNAs and miRNA mimics in turn may downregulate WNT inhibitors and upregulate WNT signaling, which then would lead to new bone formation and a decrease in bone resorption.
A number of miRNAs expressed in periodontal tissues, including miR-34a, miR-125a, miR-146a, miR-223, and miR-503, inhibit osteoclast differentiation and function by modulating various regulatory members of osteoclast differentiation signal pathways (Sugatani and Hruska 2007; Nakasa et al. 2011; Chen, Cheng, et al. 2014; Guo et al. 2014; Krzeszinskia et al. 2014). The expression of these miRNAs was downregulated during osteoclastogenesis, and this process facilitated osteoclast differentiation. This group of miRNAs targets osteoclast differentiation pathway members such as Tgif2 (miR-34a; Krzeszinskia et al. 2014); TNFR-associated factor TRAF6 (miR-125a; Guo et al. 2014); RANK (miR-503; Chen, Cheng, et al. 2014); the IL1R-associated kinases IRAK1, IRAK2, and TRAF6 (miR-146a; Pauley et al. 2008; Hou et al. 2009; Nakasa et al. 2011); and finally IKK-a and NFIA (miR-223; Li et al. 2012; Xie et al. 2015). Some of these miRNAs, such as miR-125a or miR-146a, would be ideal candidates to halt the progress of osteoclast differentiation and ameliorate periodontal disease and its effect on bone loss.
A second group of miRNAs inhibits osteoclast differentiation. The expression of these miRNAs was upregulated during osteoclastogenesis to provide negative feedback loops related to osteoclast differentiation. Among these, miR-26a affects osteoclast formation, actin-ring formation, and bone resorption by targeting the connective tissue growth factor/CCN family 2 (CTGF/CCN2) (Kim et al. 2015), while miR-155 inhibits osteoclast differentiation by inhibiting the expression of SOCS1 and MITF, 2 essential regulators of osteoclastogenesis (Mann et al. 2010; Zhang J et al. 2012). Both miR-26a and miR-155 may constitute a potential internal feedback mechanism for the control of bone remodeling. In light of their ability to inhibit osteoclastogenesis, overexpression or application of miR-26a and miR-155 mimics may also be used in future strategies for the treatment of alveolar bone loss.
Several miRNAs expressed in periodontal tissues (Table 2), including miR-21, miR-29 family, miR-31, and miR-148, directly promote osteoclast differentiation and function (Sugatani et al. 2011; Cheng et al. 2013; Franceschetti et al. 2013; Mizoguchi et al. 2013). The expression of these miRNAs was upregulated during osteoclastogenesis (Sugatani et al. 2011; Cheng et al. 2013; Franceschetti et al. 2013; Mizoguchi et al. 2013). miR-21 is a possible mediator in the proposed relationship between estrogen deficiency and periodontal disease, resulting in increased osteoclast activity by targeting Fas ligand (FASL; Sugatani and Hruska 2013) in periodontal disease patients during menopause (Shapiro and Freeman 2014). The expression of all miR-29 family members, miR-29a, miR-29b, and miR-29c, increases osteoclast differentiation, possibly through the cell division control protein 42 (Cdc42) and its effect on cytoskeleton organization (Franceschetti et al. 2013). Another miRNA that regulates osteoclast activity through the control of cytoskeletal organization and RhoA is miR-31 (Mizoguchi et al. 2013). In contrast, miR-148a regulates osteoclastogenesis by targeting the leucine zinc finger transcription factor MAFB (Cheng et al. 2013).
Therapeutic Opportunities and Challenges
This review has listed only a small fraction of the miRNAs involved in periodontal homeostasis and periodontal disease progression, and based on the estimate that miRNAs make up >4% of all human genes, it is likely that the number of these small but powerful regulators involved in the biology of the periodontium is only going to increase. miRNAs are unique regulatory molecules because of their ability to simultaneously affect sets of multiple related genes and their limited requirements for complementarity with target mRNAs (Diekwisch 2016b). Moreover, miRNAs in general do not cause drastic changes in the expression of a single gene but rather control the fine-tuning of gene expression and reduce expression noise (Schmiedel et al. 2015). As a result, miRNAs not only are of experimental or diagnostic value but also hold great potential for the treatment of disease, including periodontal disease. As therapeutics, individual miRNAs can target several genes and affect multiple regulatory networks, while a combination of miRNAs or their antagonists may be used to regulate several members of the same signaling pathway. For example, miRNA therapeutics may be applied to restore the function of lost or downregulated miRNAs related to osteoblastogenesis or to inhibit the function of upregulated miRNAs related to osteoclast differentiation and function (Fig. 4).
In recent years, more than a dozen different miRNA delivery systems have been developed, including viral and nonviral approaches (Zhang et al. 2013; Yang et al. 2015). Viral miRNA delivery systems are based on retroviruses, lentiviruses, and adenoviruses and are distinguished by a high infection efficiency and high miRNA or antagomir expression levels, while suffering from relatively higher toxicity and immunogenicity levels (Zhang et al. 2013; Yang et al. 2015; Mishra et al. 2016). In contrast, nonviral approaches face the challenge of having to transport the miRNA or its antagonist across the cell membrane and to protect it from degradation on its way to the nucleus (Zhang Z et al. 2012). These nonviral miRNA delivery approaches include lipid-based delivery systems such as liposomes as well as polymer-based approaches including polyethylenimine (PEI), poly(lactide-co-glycolide) (PLGA), and poly(amidoamine) (PAMAM) dendrimers (Zhang et al. 2013; Yang et al. 2015). Other recently developed miRNA carriers include chitosan, protamine, and collagen (Zhang et al. 2013), as well as gold-, iron-, and silica-based nanoparticles (Gaharwar et al. 2014; Xavier et al. 2015).
The ability of miRNAs to regulate multiple target genes and tissues has caused concerns about off-target effects (OTEs). To address these concerns, computational algorithms to predict, detect, and suppress OTEs in large-scale RNAi screens have been developed (Yilmazel et al. 2014; Zhong et al. 2014). While miRNAs are known to be conserved among species, OTEs are fairly species specific or at least conserved within mammalian orders, suggesting the importance of an inclusion of primate models prior to clinical trials in humans (Jackson and Linsley 2010). Novel aptamer-miRNA conjugates have been developed to deliver miRNAs into specific cell types, resulting in a reduction of the dose required for pharmacological effects, a reduction in OTEs, and reduced toxicity (Esposito et al. 2014). Most recently, a novel plasmid-based miRNA inhibitor system (PMIS) has been developed that relies on specific binding of a PMIS molecule to target miRNAs, resulting in the introduction of a new secondary structure and the formation of a stable PMIS-miR complex (Cao et al. 2016; Diekwisch 2016b).
Supplementary Material
Acknowledgments
Generous funding by National Institute of Dental and Craniofacial Research grant DE019463 (Epigenetics of Dental Stem Cells) to X.L. is gratefully acknowledged.
Footnotes
A supplemental appendix to this article is available online.
Statement of Authorship: X. Luan, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; X. Zhou, J. Trombetta-eSilva, contributed to conception, design, and data acquisition, drafted and critically revised the manuscript; M. Francis, contributed to conception, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; A.K. Gaharwar, contributed to conception and data interpretation, drafted and critically revised the manuscript; P. Atsawasuwan, contributed to design and data acquisition, drafted and critically revised the manuscript; T.G.H. Diekwisch, contributed to conception, design, data analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, Dawson BC, Munivez E, Tao J, Lee BH. 2012. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet. 21(13):2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- Bartold PM. 1995. Turnover in periodontal connective tissues: dynamic homeostasis of cells, collagen and ground substances. Oral Dis. 1(4):238–253. [DOI] [PubMed] [Google Scholar]
- Bartold PM, Van Dyke TE. 2013. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 62(1):203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce BF. 2013. Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J Bone Miner Res. 28(4):711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce BF, Xiu Y, Li J, Xing L, Yao Z. 2015. NF-κB-mediated regulation of osteoclastogenesis. Endocrinol Metab (Seoul). 30(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310(5747):486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Yu W, Li X, Wang J, Gao S, Holton NE, Eliason S, Sharp T, Amendt BA. 2016. A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther. 23(7):634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell. 136(4):642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 157(1):77–94. [DOI] [PubMed] [Google Scholar]
- Cekici A, Kantarci A, Hasturk H, Van Dyke TE. 2014. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000. 64(1):57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng P, Xie H, Zhou HD, Wu XP, Liao EY, Luo XH. 2014. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res. 29(2):338–347. [DOI] [PubMed] [Google Scholar]
- Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi K, Hokland L, Kassem M. 2014. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 32(4):902–912. [DOI] [PubMed] [Google Scholar]
- Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, et al. 2013. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. 28(5):1180–1190. [DOI] [PubMed] [Google Scholar]
- Dangaria SJ, Ito Y, Luan X, Diekwisch TG. 2011a. Successful periodontal ligament regeneration by periodontal progenitor preseeding on natural tooth root surfaces. Stem Cells Dev. 20(10):1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangaria SJ, Ito Y, Luan X, Diekwisch TGH. 2011b. Differentiation of neural-crest-derived intermediate pluripotent progenitors into committed periodontal populations involves unique molecular signature changes, cohort shifts, and epigenetic modifications. Stem Cells Dev. 20(1):39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangaria SJ, Ito Y, Walker C, Druzinsky R, Luan X, Diekwisch TGH. 2009. Extracellular matrix-mediated differentiation of periodontal progenitor cells. Differentiation. 78(2–3):79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangaria SJ, Ito Y, Yin LL, Valdré G, Luan X, Diekwisch TGH. 2011. Apatite microtopographies instruct signaling tapestries for progenitor-driven new attachment of teeth. Tissue Eng Part A. 17(3–4):279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8(7):481–490. [DOI] [PubMed] [Google Scholar]
- Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y, Gu P, Fan X. 2013. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 22(16):2278–2286. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG. 2002. Pathways and fate of migratory cells during late tooth organogenesis. Connect Tissue Res. 43(2–3):245–256. [PubMed] [Google Scholar]
- Diekwisch TG. 2016a. Our periodontal tissue: a masterpiece of evolution. J Clin Periodontol. 43(4):320–322. [DOI] [PubMed] [Google Scholar]
- Diekwisch TGH. 2016b. Novel approaches toward managing the micromanagers: ‘non-toxic’ but effective. Gene Therapy. 23(10):697–698. [DOI] [PubMed] [Google Scholar]
- Esposito CL, Cerchia L, Catuogno S, De Vita G, Dassie JP, Santamaria G, Swiderski P, Condorelli G, Giangrande PH, de Franciscis V. 2014. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol Ther. 22(6):1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschetti T, Kessler CB, Lee SK, Delany AM. 2013. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem. 288(46):33347–33360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaharwar AK, Peppas NA, Khademhosseini A. 2014. Nanocomposite hydrogels for biomedical applications. Biotechnol Bioeng. 111(3):441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Zheng L. 2013. microRNA expression in rat apical periodontitis bone lesion. Bone Res. 1(2):170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay I, Cavender A, Peto D, Sun Z, Speer A, Cao H, Amendt BA. 2014. Differentiation of human dental stem cells reveals a role for microRNA-218. J Periodont Res. 49(1):110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves D. 2008. Cytokines that promote periodontal tissue destruction. J Periodontol. 79(Suppl 8):1585–1591. [DOI] [PubMed] [Google Scholar]
- Graves DT, Li J, Cochran DL. 2011. Inflammation and uncoupling as mechanisms of periodontal bone loss. J Dent Res. 90(2):143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünhagen J, Bhushan R, Degenkolbe E, Jäger M, Knaus P, Mundlos S, Robinson PN, Ott CE. 2015. MiR-497∼195 cluster microRNAs regulate osteoblast differentiation by targeting BMP signaling. J Bone Miner Res. 30(5):796–808. [DOI] [PubMed] [Google Scholar]
- Guo LJ, Liao L, Yang L, Li Y, Jiang TJ. 2014. MiR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp Cell Res. 321(2):142–152. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15(8):509–524. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hienz SA, Paliwal S, Ivanovski S. 2015. Mechanisms of bone resorption in periodontitis. J Immunol Res. 2015:615486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. 2007. How microRNAs choose their targets. Nat Genet. 39(10):1191–1192. [DOI] [PubMed] [Google Scholar]
- Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. 2009. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 183(3):2150–2158. [DOI] [PubMed] [Google Scholar]
- Hung PS, Chen FC, Kuang SH, Kao SY, Lin SC, Chang KW. 2010. miR-146a induces differentiation of periodontal ligament cells. J Dent Res. 89(3):252–257. [DOI] [PubMed] [Google Scholar]
- Irwandi RA, Vacharaksa A. 2016. The role of microRNA in periodontal tissue: a review of the literature. Arch Oral Biol. 72:66–74. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 9(1):57–67. [DOI] [PubMed] [Google Scholar]
- Jeong BC, Kang IH, Hwang YC, Kim SH, Koh JT. 2014. MicroRNA-194 reciprocally stimulates osteogenesis and inhibits adipogenesis via regulating COUP-TFII expression. Cell Death Dis. 5:e1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu X, Feng Y, Dai Z. 2014. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp Mol Med. 46:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung IH, Yun JH, Cho AR, Kim CS, Chung WG, Choi SH. 2011. Effect of (–)-epigallocatechin-3-gallate on maintaining the periodontal ligament cell viability of avulsed teeth: a preliminary study. J Periodontal Implant Sci. 41(1):10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalea AZ, Hoteit R, Suvan J, Lovering RC, Palmen J, Cooper JA, Khodiyar VK, Harrington Z, Humphries SE, D’Aiuto F. 2015. Upregulation of gingival tissue miR-200b in obese periodontitis subjects. J Dent Res. 94(Suppl 3):59S–69S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama Y. 1973. The pattern of alveolar bone activity during development and eruption of the molar in the rat. J Periodontal Res. 8(3):179–191. [DOI] [PubMed] [Google Scholar]
- Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. 2010. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 285(33):25221–25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 24:389–406. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim JH, Kim I, Lee J, Seong S, Park YW, Kim N. 2015. MicroRNA-26a regulates RANKL-induced osteoclast formation. Mol Cells. 38(1):75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeszinskia JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et al. 2014. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 512(7515):431–435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 21(3):172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Song DH, Kim YJ, Choi B, Chung YH, Kim SM, Koh JM, Yoon SY, Song Y, Kang SW, et al. 2014. PTX3 stimulates osteoclastogenesis by increasing osteoblast RANKL production. J Cell Physiol. 229(11):1744–1752. [DOI] [PubMed] [Google Scholar]
- Lee YH, Na HS, Jeong SY, Jeong SH, Park HR, Chung J. 2011. Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. Biocell. 35(2):43–49. [PubMed] [Google Scholar]
- Li YT, Chen SY, Wang CR, Liu MF, Lin CC, Jou IM, Shiau AL, Wu CL. 2012. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 64(10):3240–3245. [DOI] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2009. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 284(23):15676–15684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. 2012. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 8(4):212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WH, Liu B, Cheng D, Williams BO, Mah SJ, Helms JA. 2014. Wnt signaling regulates homeostasis of the periodontal ligament. J Periodontal Res. 49(6):751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WH, Liu B, Mah SJ, Yin X, Helms JA. 2015. Alveolar bone turnover and periodontal ligament width are controlled by Wnt. J Periodontol. 86(2):319–326. [DOI] [PubMed] [Google Scholar]
- Lin GJ, Hankenson KD. 2011. Integration of BMP, Wnt, and Notch signaling pathways in osteoblast differentiation. J Cell Biochem. 112(12):3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu W, Hu C, Xue Z, Wang G, Ding B, Luo H, Tang L, Kong X, Chen X, et al. 2011. MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells. 29(11):1804–1816. [DOI] [PubMed] [Google Scholar]
- Löe H, Theilade E, Jensen SB. 1965. Experimental gingivitis in man. J Periodontol. 36:177–187. [DOI] [PubMed] [Google Scholar]
- Luan X, Dangaria S, Ito Y, Walker CG, Jin T, Schmidt MK, Galang MT, Druzinsky R. 2009. Neural crest lineage segregation a blueprint for periodontal regeneration. J Dent Res. 88(9):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X, Ito Y, Dangaria S, Diekwisch TG. 2006. Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cells Dev. 15(4):595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane LA, Murphy PR. 2010. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 11(7):537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Barad O, Agami R, Geiger B, Hornstein E. 2010. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci U S A. 107(36):15804–15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. 2006. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 13(12):1102–1107. [DOI] [PubMed] [Google Scholar]
- Marsh PD, Devine DA. 2011. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 38(Suppl 11):28–35. [DOI] [PubMed] [Google Scholar]
- McIntosh JE, Anderton X, FloresDeJacoby L, Carlson DS, Shuler CF, Diekwisch TG. 2002. Caiman periodontium as an intermediate between basal vertebrate ankyloses-type attachment and mammalian “true” periodontium. Microsc Res Tech. 59(5):449–459. [DOI] [PubMed] [Google Scholar]
- Millar NL, Gilchrist DS, Akbar M, Reilly JH, Kerr SC, Campbell AL, Murrell GA, Liew FY, Kurowska-Stolarska M, McInnes IB. 2015. MicroRNA29a regulates IL-33-mediated tissue remodelling in tendon disease. Nat Commun. 6:6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Yadav T, Rani V. 2016. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 98:12–23. [DOI] [PubMed] [Google Scholar]
- Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M. 2010. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. 109(5):866–875. [DOI] [PubMed] [Google Scholar]
- Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. 2013. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res Ther. 15(5):R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri T, Hanada K, Ozawa H. 1991. Coupling of resorption and formation on bone remodeling sequence in orthodontic tooth movement: a histochemical study. J Bone Miner Metab. 9(2):57–69. [Google Scholar]
- Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. 2011. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 63(6):1582–1590. [DOI] [PubMed] [Google Scholar]
- Ogata Y, Matsui S, Kato A, Zhou L, Nakayama Y, Takai H. 2014. MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients. J Oral Sci. 56(4):253–260. [DOI] [PubMed] [Google Scholar]
- Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. 2014. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014:970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios S, Xiao W, Mattos M, Lim J, Tarapore RS, Alsadun S, Yu B, Wang CY, Graves D. 2015. Osteoblast lineage cells play an essential role in periodontal bone loss through activation of nuclear factor-kappa B. Scientific Reports. 5:16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC, Schroeder HE. 1976. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab Invest. 34(3):235–249. [PubMed] [Google Scholar]
- Park MG, Kim JS, Park SY, Lee SA, Kim HJ, Kim CS, Kim JS, Chun HS, Park JC, Kim DK. 2014. MicroRNA-27 promotes the differentiation of odontoblastic cell by targeting APC and activating Wnt/β-catenin signaling. Gene. 538(2):266–272. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. 2012. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 13(4):271–282. [DOI] [PubMed] [Google Scholar]
- Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. 2008. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 10(4):R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri R, Nares S, Zhang S, Barros SP, Offenbacher S. 2012. MicroRNA modulation in obesity and periodontitis. J Dent Res. 91(1):33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich K, Smolen JS. 2012. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 11(3):234–250. [DOI] [PubMed] [Google Scholar]
- Rippin JW. 1976. Collagen turnover in the periodontal ligament under normal and altered functional forces. I. Young rat molars. J Periodontal Res. 11(2):101–107. [DOI] [PubMed] [Google Scholar]
- Rippin JW. 1978. Collagen turnover in the periodontal ligament under normal and altered functional forces. II. Adult rat molars. Periodontal Res. 13(2):149–154. [DOI] [PubMed] [Google Scholar]
- Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Blüthgen N, Marks DS, van Oudenaarden A. 2015. Gene expression: microRNA control of protein expression noise. Science. 348(6230):128–132. [DOI] [PubMed] [Google Scholar]
- Shapiro LF, Freeman K. 2014. The relationship between estrogen, estrogen receptors and periodontal disease in adult women: a review of the literature. N Y State Dent J. 80(3):30–34. [PubMed] [Google Scholar]
- Sodek J, Ferrier JM. 1988. Collagen remodelling in rat periodontal tissues: compensation for precursor reutilization confirms rapid turnover of collagen. Coll Relat Res. 8(1):11–21. [DOI] [PubMed] [Google Scholar]
- Song Q, Zhong L, Chen C, Tang Z, Liu H, Zhou Y, Tang M, Zhou L, Zuo G, Luo J, et al. 2015. miR-21 synergizes with BMP9 in osteogenic differentiation by activating the BMP9/Smad signaling pathway in murine multilineage cells. Int J Mol Med. 36(6):1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. 2012. MicroRNAs and their target genes in gingival tissues. J Dent Res. 91(10): 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. 2007. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 101(4):996–999. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. 2013. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. 114(6):1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Vacher J, Hruska KA. 2011. A microRNA expression signature of osteoclastogenesis. Blood. 117(13):3648–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M, Nemoto E. 2016. Role of the Wnt signaling molecules in the tooth. Japanese Dent Sci Rev. 52(4):75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL. 2000. Bone resorption by osteoclasts. Science. 289(5484):1504–1508. [DOI] [PubMed] [Google Scholar]
- Theilade E, Wright WH, Jensen SB, Löe H. 1966. Experimental gingivitis in man. II. A longitudinal clinical and bacteriological investigation. J Periodontal Res. 1(1):1–13. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20(5):515–524. [DOI] [PubMed] [Google Scholar]
- Vimalraj S, Partridge NC, Selvamurugan N. 2014. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 229(9):1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhou Z, Wu T, Liu W, Yin P, Pan C, Yu X. 2016. TNF-α-induced NF-κB activation upregulates microRNA-150-3p and inhibits osteogenesis of mesenchymal stem cells by targeting β-catenin. Open Biol. 6(3):pii:150258. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang T, Xu Z. 2010. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. 402(2):186–189. [DOI] [PubMed] [Google Scholar]
- Wiebe SH, Hafezi M, Sandhu HS, Sims SM, Dixon SJ. 1996. Osteoclast activation in inflammatory periodontal diseases. Oral Dis. 2(2):167–180. [DOI] [PubMed] [Google Scholar]
- Wu T, Xie M, Wang X, Jiang X, Li J, Huang H. 2012. miR-155 modulates TNF-α-inhibited osteogenic differentiation by targeting SOCS1 expression. Bone. 51(3):498–505. [DOI] [PubMed] [Google Scholar]
- Xavier JR, Thakur T, Desai P, Jaiswal MK, Sears N, CosgriffHernandez E, Kaunas R, Gaharwar AK. 2015. Bioactive nanoengineered hydrogels for bone tissue engineering: a growth-factor-free approach. ACS Nano. 9(3):3109–3118. [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P, Fan X. 2014. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun. 446(1):98–104. [DOI] [PubMed] [Google Scholar]
- Xie Y, Zhang L, Gao Y, Ge W, Tang P. 2015. The multiple roles of microrna-223 in regulating bone metabolism. Molecules. 20(10):19433–19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL. 2011. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. Int J Oral Sci. 3(3):125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, Yin VP, Lockman P, Bai S. 2015. Exosome delivered anticancer drugs across the bloodbrain barrier for brain cancer therapy in Danio rerio. Pharm Res. 32(6):2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmazel B, Hu Y, Sigoillot F, Smith JA, Shamu CE, Perrimon N, Mohr SE. 2014. Onlin GESS: prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinformatics. 15:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Li J, Salmon B, Huang L, Lim WH, Liu B, Hunter DJ, Ransom RC, Singh G, Gillette M, et al. 2015. Wnt signaling and its contribution to craniofacial tissue homeostasis. J Dent Res. 94(11):1487–1494. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q, Lin R, Han Q, Li J, Zhao RC. 2012. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 586(16):2375–2381. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, Shen J, Huang Y. 2012. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. 586(19): 3255–3262. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Gemeinhart RA. 2013. Progress in microRNA delivery. J Control Release. 172(3):962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. 2011. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 108(24):9863–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qin YW, Brewer G, Jing Q. 2012. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip Rev RNA. 3(4):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Li Y, Lin Z, Wan J, Xu C, Zeng Y, Zhu Y. 2016. miR-199b-5p modulates BMSC osteogenesis via suppressing GSK-3β/β-catenin signaling pathway. Biochem Biophys Res Commun. 477(4):749–754. [DOI] [PubMed] [Google Scholar]
- Zhong R, Kim J, Kim HS, Kim M, Lum L, Levine B, Xiao G, White MA, Xie Y. 2014. Computational detection and suppression of sequence-specific off-target phenotypes from whole genome RNAi screens. Nucleic Acids Res. 42(13):8214–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Luan X, Chen Z, Francis M, Gopinathan G, Li W, Lu X, Li S, Wu C, Diekwisch TG. 2016. MicroRNA-138 inhibits periodontal progenitor differentiation under inflammatory conditions. J Dent Res. 95(2):230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.