Figure 2.
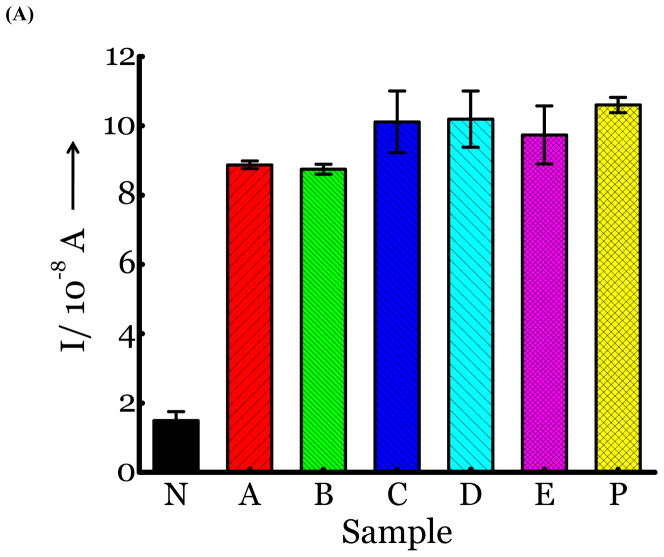
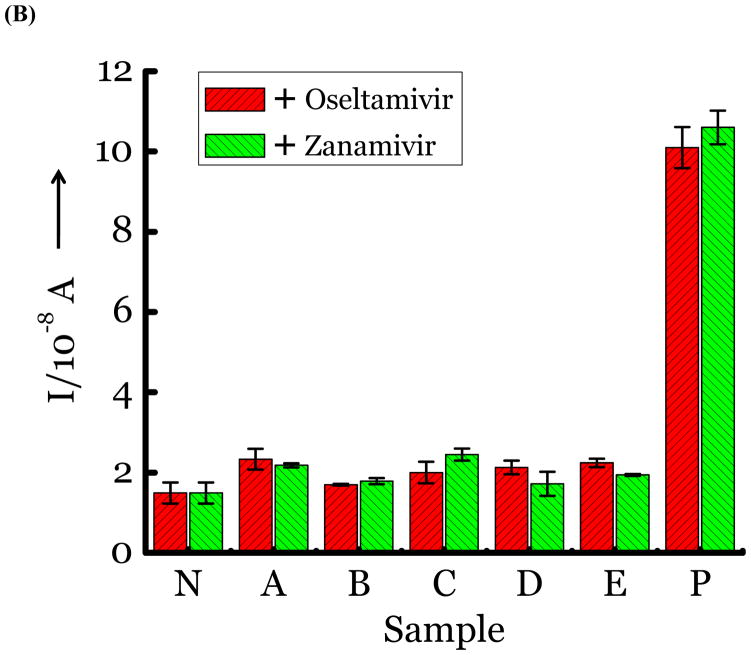
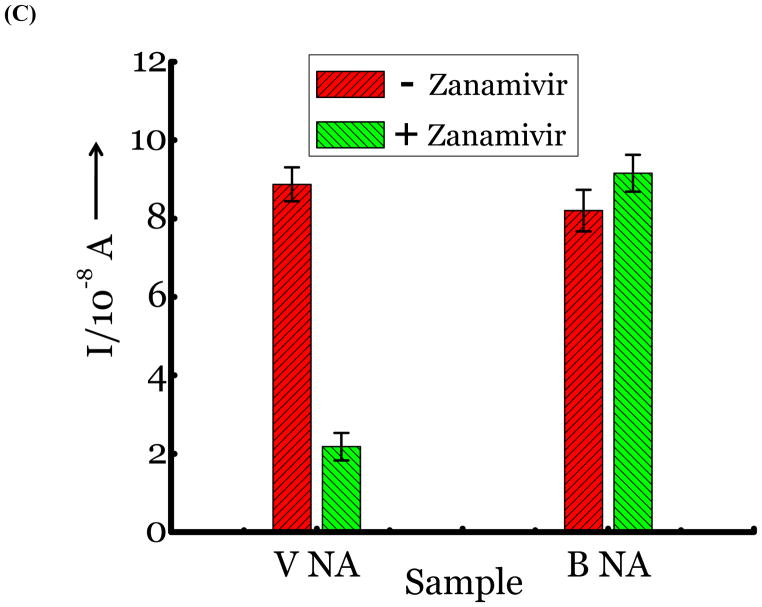
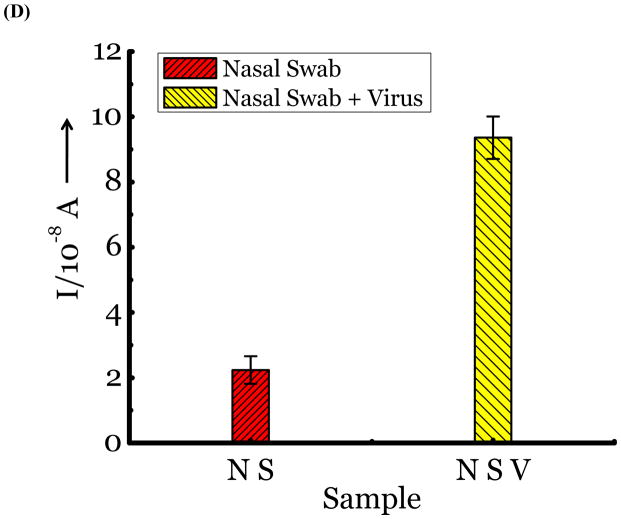
(A) Detection of Influenza virus or viral NA. SG1 (0.5 mM) was incubated with membrane free soluble N1 NA (Sample A, strain H5N1 A/Anhui/1/2005) or N2 NA (Sample B, strain H3N2 A/Babol/36/2005) or three different UV inactivated influenza strains, H3N2 A/Aichi/2/1968 (Sample C) or H1N1 A/Brisbane/59/2007 (Sample D), H3N2 A/HongKong/8/68 (Sample E) for two hours. Glucose was released in all samples. The negative control where no virus or NA was added (Sample N) did not show any noticeable current and the positive control was D-glucose (Sample P) at 0.5 mM. (B) Drug Susceptibility Studies. 10 ng of FDA approved antiviral Zanamivir or Oseltamivir (Carbosynth, USA, San Diego, CA) Zanamivir® or Oseltamivir® were premixed with the strains for 30 min at rt before addition of SG1. Glucose was not released because these strains are susceptible to the drugs. (C) Studies with bacterial NA. Bacterial NA cleaves SG1 to release glucose, however, Zanamivir does not inhibit bacterial NA and glucose is released when BNA was premixed with Zanamivir and incubated with SG1. (D) Studies with human samples. A. Nasal Swab only. There is no glucose present. B. Nasal swab spiked with 105 pfu of H1N1 A/Brisbane/59/2007 and added to SG1. The positive signal indicates there are no matrix effects. The y axis represents current, I, in amperes measured after 100 s using an amperometric i-t curve at a working potential of 0.00 V, and the x-axis represents different samples. All experiments were performed in triplicate independently on different days to demonstrate robustness of the assay.