Abstract
Prader-Willi syndrome is a neurodevelopmental disorder that is characterized by infantile hypotonia, feeding difficulties, hypogonadism, mental deficiency, hyperphagia (leading to obesity in early childhood), learning problems, and behavioral difficulties. A paternal 15q11-q13 deletion is found in ~70% of patients with Prader-Willi syndrome, ~25% have uniparental maternal disomy 15, and the remaining 2% to 5% have imprinting defects. The proximal deletion breakpoint in the 15q11-q13 region occurs at 1 of 2 sites located within either of 2 large duplicons allowing for the identification of 2 deletion subgroups. The larger, type I (TI) deletion involves breakpoint 1, which is close to the centromere, whereas the smaller, type II (TII) deletion involves breakpoint 2, located ~500 kilobases distal to breakpoint 1. Breakpoint 3 is located at the distal end of the 15q11-q13 region and common to both typical deletion subgroups. Analyses of the genetic subtypes of Prader-Willi syndrome to date have primarily compared individuals with typical deletion and uniparental maternal disomy 15 without grouping the individuals with a deletion into TI or TII. Distinct differences have been reported between individuals with Prader-Willi syndrome resulting from deletion compared with uniparental maternal disomy 15 in physical, cognitive, and behavioral parameters. We previously presented the first assessment of clinical differences in individuals with Prader-Willi syndrome categorized as having type I or II deletions. Adaptive behavior, obsessive-compulsive behaviors, reading, math, and visual-motor integration assessments were generally poorer in individuals with Prader-Willi syndrome and the TI deletion compared with subjects with Prader-Willi syndrome with the TII deletion or uniparental maternal disomy 15. Four genes (NIPA1, NIPA2, CYFIP1, and GCP5) have been identified in the chromosomal region between breakpoints 1 and 2 and are implicated in compulsive behavior and lower intellectual ability observed in individuals with Prader-Willi syndrome with TI versus TII deletions. We quantified messenger-RNA levels of these 4 genes in actively growing lymphoblastoid cells derived from 8 subjects with Prader-Willi syndrome with the TI deletion (4 males, 4 females; mean: age 25.2 ± 8.9 years) and 9 with the TII deletion (3 males, 6 females; mean age: 19.5 ± 5.8 years). Messenger-RNA levels were correlated with validated psychological and behavioral scales administered by trained psychologists blinded to genotype status. Messenger RNA from NIPA1, NIPA2, CYFIP1, and GCP5 was reduced but detectable in the subjects with Prader-Willi syndrome with the TI deletion, supporting biallelic expression. For the most part, messenger-RNA values were positively correlated with assessment parameters, indicating a direct relationship between messenger-RNA levels and better assessment scores, with the highest correlation for NIPA2. The coefficient of determination indicated the quantity of messenger RNA of the 4 genes explained from 24% to 99% of the variation of the behavioral and academic parameters measured. By comparison, the co-efficient of determination for deletion type alone explained 5% to 50% of the variation in the assessed parameters. Understanding the influence of gene expression on behavioral and cognitive characteristics in humans is in the early stage of research development. Additional research is needed to identify the function of these genes and their interaction with gene networks to clarify the potential role they play in central nervous system development and function.
Keywords: Prader-Willi syndrome, PWS, type I and II deletions, genotype-phenotype correlation, academic and behavioral assessment, gene expression
Prader-willi syndrome (PWS) is a neurodevelopmental disorder resulting from loss of function of paternally expressed genes from the 15q11-q13 region and characterized by severe hypotonia, feeding difficulties, hypogonadism, small hands and feet, developmental delay, behavior problems, and genital hypoplasia. However, children with PWS between 2 and 4 years of age develop an insatiable appetite and become severely obese if food intake is not strictly controlled.1–3
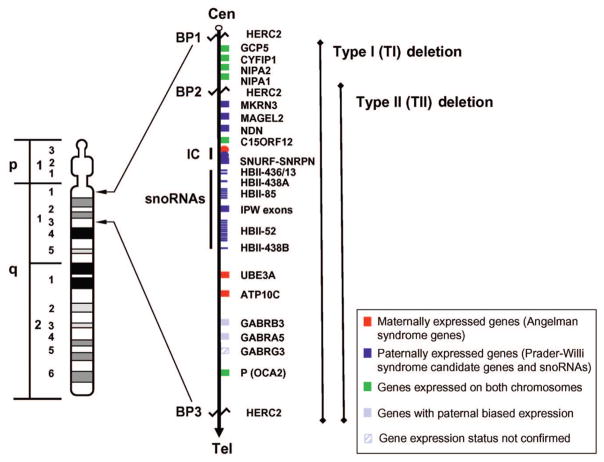
A paternal 15q11-q13 deletion is found in ~70% of subjects with PWS, ~25% have uniparental maternal disomy 15 (UPD), and the remaining 2% to 5% have imprinting defects.3,4 The proximal deletion breakpoint (BP) in the 15q11-q13 region occurs at 1 of 2 sites located within either of 2 large duplicons centromeric to locus ZNF127,5,6 which allows for the identification of 2 deletion subgroups. The larger type I (TI) deletion involves BP1, which is close to the centromere, and the smaller type II (TII) deletion involves BP2, located ~500 kilobase (kb) distal to BP1 (see Fig 1). BP3 is located at the distal end of the 15q11-q13 region and common to both deletion subgroups.
FIGURE 1.
High-resolution chromosome 15 ideogram, order of genes on 15q11-q13 region, and patterns of expression. Gene order is shown according to the UCSC Genome Bioinformatics site (http://genome.ucsc.edu). snoRNA indicates small nucleolar RNA. (Adapted from Bittel DC, Bulter MG. Expert Rev Mol Med. 2005;7:1–20.)
Analyses of the genetic subtypes of PWS to date have primarily compared typical individuals with deletion and UPD without grouping the individuals with a deletion into TI or TII. For example, hypopigmentation and homogeneous clinical presentations are more often seen in individuals with PWS with a deletion compared with those having normal chromosomes now recognized with UPD or an imprinting defect.1,7,8 Significantly higher verbal IQ scores in 4 subcategories of verbal testing (information, arithmetic, vocabulary, and comprehension) have been reported in individuals with PWS with UPD compared to individuals with a deletion.9 In addition, individuals with PWS with UPD have been reported to have fewer maladaptive behaviors and reduced self-injury compared with individuals with deletions.10–12 Conversely, visual processing of complex stimuli was significantly poorer in individuals with UPD compared with those with the deletion.13
We previously presented the first assessment of clinical differences in individuals with PWS who were categorized as having TI or TII deletions14 using a large existing clinical and behavioral data set. Significant differences were found between the 2 typical deletion groups and those with UPD. For example, adaptive-behavior scores were generally worse in individuals with PWS and the TI deletion, as were specific obsessive-compulsive behaviors compared with subjects with PWS resulting from UPD.14 Individuals with PWS with TI deletions had poorer reading and math skills and visual-motor integration. In general, individuals with TI deletions had more behavioral and psychological problems than individuals with the TII deletion or UPD.
Two recent reports have also evaluated the possible contribution of chromosome 15 genetic subtypes to clinical presentation of individuals with PWS.15,16 These 2 studies reported a trend toward poorer performance in subjects with PWS with TI deletions compared with subjects with TII deletions based on parental reports of behavioral and intellectual observations particularly associated with autism.
Four genes (NIPA1, NIPA2, CYFIP1, and GCP5) have been identified in the chromosomal region between BP1 and BP25 and are implicated in compulsive behavior and lower intellectual ability in individuals with PWS with TI versus TII deletions. NIPA1 is expressed in mouse brain tissue but is not imprinted. In addition, NIPA1 is implicated in spastic paraplegia,17 suggesting that it may be important for central nervous system development and/or function. Hence, we report gene-expression studies with the 4 genes located between BP1 and BP2 in those individuals with PWS with TI or TII deletions previously studied14 and examined the relationship between messenger-RNA (mRNA) levels and behavioral-and intellectual-assessment scores.
METHODS
Our PWS study subjects included 8 subjects with TI deletions (4 males, 4 females; mean age: 25.2 ± 8.9 years) and 9 with TII deletions (3 males, 6 females; mean age: 19.5 ± 5.8 years). The PWS diagnosis was confirmed by methylation testing, and the deletion status was established by fluorescence in situ hybridization. By genotyping informative microsatellites (eg, D15S1035) between BP1 and BP2, the paternal deletion in this region was identified in the subjects with TI deletions as described previously.14 These individuals represent a subset of the group we previously reported14 for whom we were able to generate lymphoblastoid cell cultures for isolation of RNA to allow quantification of mRNA.
Several validated psychological and behavioral scales were administered by trained psychologists blinded to genotype status to assess phenotypic characteristics of individuals with PWS as previously described.14 These scales included the Yale-Brown Obsessive Compulsive Scale,18,19 used to measure obsessions and compulsions in psychiatric patients including individuals with PWS20,21; the Compulsive Behavior Checklist (CBC),22 designed for people with intellectual disabilities focusing on compulsive behavior rather than obsessions; the Reiss Screen for Maladaptive Behavior,23 to assess psychiatric symptoms of people with developmental disabilities; the Scales of Independent Behavior (SIB),24 to assess both adaptive and maladaptive behavior of individuals with cognitive disabilities; the Wechsler Intelligence Scale, to evaluate intellectual ability25,26; and the Visual Motor Integrations (VMI) Scale27 to measure visual-motor skills. Academic skills were assessed by using the Woodcock-Johnson Psycho-Educational Battery–Revised.28 Not all subjects were cooperative; therefore, there are missing data for some assessments. Statistical analyses included t test, correlation, and coefficient of determination.
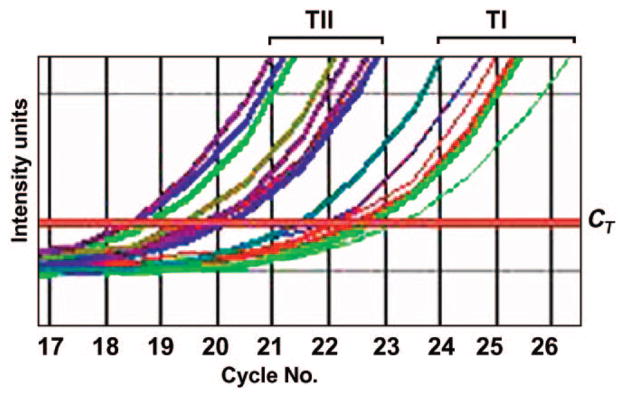
Quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed by using a QuantiTect SYBR green RT-PCR kit (Qiagen, Valencia, CA) according to manufacturer directions. Total RNA was isolated from cell lines by using Trizol reagent (Invitrogen, Inc, Carlsbad, CA) and quantified by spectroscopy. Enough RNA was isolated in a single round of purification to carry out all RT-PCRs. An equal quantity of total RNA (500 ng) from each subject together with primers specific to the gene being quantified were added to a reaction mix containing all components necessary for RT and PCR. The reaction was conducted in an ABI 7000 system (Applied Biosystems, Foster City, CA) beginning with a 30-minute step at 50°C to allow for RT followed by 15 minutes at 95°C. PCR was performed for 45 cycles during which the intensity of the SYBR green fluorescence was measured at the extension step of each PCR cycle. The point at which the intensity level crossed the PCR cycle threshold (CT, defined as the narrowest point between individual reactions in the logarithmic phase of the reaction) was used to compare individual reactions (see Fig 2). At least 5 replicates were performed on each sample for each gene. A dissociation curve was generated for all reactions, and reactions were run on agarose gels to verify the presence of a single band. Quantitative RT-PCR was also performed by using primers specific to GAPD, a control gene, on all RNA samples. All samples were normalized individually to GAPD expression by dividing the mean GAPD gene expression (CT) value from each subject to the mean GAPD gene expression (CT) value of 1 of the reference subjects with PWS to produce a correction value. Each mean CT value for the other genes was divided by the correction value to produce the normalized value. The normalized CT values were averaged to produce the mean CT value for each gene analyzed.
FIGURE 2.
Representative quantitative RT-PCR analysis of NIPA2 between subjects with TI and TII PWS deletions. Samples from the subjects with TI deletions required an average of 2.5 cycles longer to reach the cycle threshold (CT) than the samples from the subjects with TII deletions. This represents an approximate 5.9-fold reduction in mRNA in the subjects with TI deletions (2|TI CT|− TII CT|=2|21.9 − 19.3| = 22.57 or 5.9).
Differences in the mean value of mRNA level were compared by t test. We evaluated CT values (explained above) between groups that were inversely related to the quantity of mRNA (ie, a larger CT value reflects a lower quantity of mRNA). SPSS software (SPSS Inc, Chicago, IL) was used to ascertain the correlation coefficients and coefficient of determination values for mRNA levels for individual genes and for their joint impact on the behavioral and intellectual assessments. The coefficient of determination is the proportion of a sample variance of a response variable that is “explained” by the predictor variables when a linear regression is performed. The evaluation of the genes in explaining the variation in phenotypic measures was investigated by fitting a general linear model and specifying quantity of mRNA (CT value) as the predictor variable and the behavioral or cognitive values as the response variable.
RESULTS
We found reduced but detectable quantities of mRNA in subjects with TI deletions compared with individuals with TII deletions, supporting biallelic expression of the 4 genes (NIPA1, NIPA2, CYFIP1, and GCP5). The mRNA from the 4 genes between BP1 and BP2 was significantly reduced in the subjects with TI deletion (Table 1), which was as expected because they have only a single copy of each gene (Fig 2). We analyzed the quantity of mRNA of each gene and compared with behavioral and psychological measures previously shown to differ between subjects with TI and TII deletions (Tables 2 and 3). The correlation coefficients are shown in Table 2. The quantity of mRNA of NIPA2 seems to have the highest level of correlation with the parameters examined. For the most part, mRNA values (which were based on relative changes in mRNA derived from CT values; Fig 2) were positively correlated with assessment parameters (Fig 3), indicating an inverse relationship with mRNA levels. We also evaluated the coefficient of determination for the genes individually and jointly (Table 3). Table also shows the contribution made by deletion type alone. For example, the coefficient of determination for the 4 genes was as low as 0.24 compared with 0.05 for deletion alone for visual-motor integration (standard score) and as high as 0.99 compared with 0.42 for deletion alone for SIB disruptive behavior severity (Table 3).
TABLE 1.
t Test Results for Subjects With TI and TII Deletions
| Gene | N | Mean CT Value | SD | Fold Change | P |
|---|---|---|---|---|---|
| NIPA1 | 4.3 | .001 | |||
| TI | 8 | 19.95 | 0.77 | ||
| TII | 9 | 17.84 | 1.30 | ||
| NIPA2 | 5.9 | .003 | |||
| TI | 8 | 21.90 | 1.86 | ||
| TII | 9 | 19.33 | 1.16 | ||
| CYFIP1 | 4.7 | .007 | |||
| TI | 8 | 24.61 | 1.77 | ||
| TII | 9 | 22.37 | 1.17 | ||
| GCP5 | 4.1 | .048 | |||
| TI | 8 | 22.32 | 2.63 | ||
| TII | 9 | 20.29 | 0.98 |
Gene-expression fold change is calculated as 2|TI CT−TII CT|; for example, the fold change between subjects with PWS with TI deletions compared to those with TII deletions for NIPA1 = 2|19.95 − 17.84| − 22.11 or 4.3.
TABLE 2.
Correlations of Gene Expression (CT Values) With Behavioral and Cognition Data Classified Into 5 Categories
| N | NIPA1 | NIPA2 | CYFIP1 | GCP5 | |
|---|---|---|---|---|---|
| Maladaptive behavior | |||||
| SIB: externalized maladaptive index | 17 | −0.23 | −0.52a | −0.47 | 0.16 |
| SIB: disruptive behavior-frequency | 17 | 0.45 | 0.55a | 0.34 | 0.24 |
| SIB: disruptive behavior-severity | 17 | 0.51a | 0.61b | 0.29 | 0.15 |
| Adaptive behavior | |||||
| SIB: broad independence score (standard score) | 17 | −0.47 | −0.62b | −0.36 | 0.10 |
| SIB: motor skills | 17 | −0.40 | −0.71c | −0.54a | 0.01 |
| SIB: social interaction and communication | 17 | −0.60a | −0.83c | −0.59a | −0.07 |
| SIB: personal living skills | 17 | −0.62b | −0.72c | −0.43 | −0.04 |
| Academic achievement | |||||
| Digit symbol WAIS-R only (standard score) | 12 | −0.32 | −0.59a | −0.18 | 0.22 |
| Woodcock-Johnson: reading cluster | 17 | −0.40 | −0.57a | −0.38 | −0.02 |
| Letter/word identification | 17 | −0.44 | −0.58a | −0.37 | −0.03 |
| Reading comprehension | 17 | −0.44 | −0.58a | −0.36 | 0.04 |
| Woodcock-Johnson: math cluster | 17 | −0.37 | −0.58a | −0.34 | −0.12 |
| Applied problems (math, standard score) | 17 | −0.45 | −0.62b | −0.39 | −0.10 |
| WRAT-3: spelling (standard score) | 17 | −0.33 | −0.53a | −0.46 | −0.20 |
| Visual-motor integration | |||||
| VMI (standard score) | 16 | −0.40 | −0.65b | −0.55a | −0.09 |
| VMI: other | 15 | −0.30 | −0.59a | −0.46 | −0.20 |
| Obsessive-compulsive behavior | |||||
| CBC: No. of categories represented | 15 | 0.42 | 0.63b | 0.41 | −0.07 |
| CBC: interference with social | 15 | 0.31 | 0.66b | 0.69b | 0.23 |
| CBC: if interrupted–halts and resumes | 15 | 0.42 | 0.37 | 0.53a | 0.14 |
| CBC: if interrupted–other | 13 | 0.29 | 0.68b | 0.33 | 0.13 |
WAIS-R indicates Wechsler Adult Intelligence Scale-Revised25; WRAT-3, Wide-Range Achievement Test, 3rd Edition. Significant Spearman’s correlation coefficient:
P < .05;
P < .01;
P < .001.
TABLE 3.
Coefficient of Determination Values for Individual Genes, Joint Impact of the 4 Genes, and Deletion Type
| NIPA1 | NIPA2 | CYFIP1 | GCP5 | Joint Impact (All 4 Genes) | Deletion Type | |
|---|---|---|---|---|---|---|
| Maladaptive behavior | ||||||
| SIB: externalized maladaptive index | 0.00 | 0.09 | 0.15 | 0.00 | 0.30 | 0.23 |
| SIB: disruptive behavior-frequency | 0.75 | 0.47 | 0.15 | 0.27 | 0.97 | 0.22 |
| SIB: disruptive behavior-severity | 0.65 | 0.40 | 0.09 | 0.04 | 0.99 | 0.42 |
| Adaptive behavior | ||||||
| SIB: broad independence score (standard score) | 0.35 | 0.20 | 0.15 | 0.04 | 0.78 | 0.12 |
| SIB: motor skills | 0.08 | 0.37 | 0.52 | 0.00 | 0.84 | 0.17 |
| SIB: social interaction and communication | 0.40 | 0.38 | 0.33 | 0.00 | 0.80 | 0.36 |
| SIB: personal living skills | 0.68 | 0.40 | 0.16 | 0.01 | 0.73 | 0.50 |
| Academic achievement | ||||||
| Digit symbol WAIS-R only (standard score) | 0.12 | 0.19 | 0.11 | 0.01 | 0.39 | 0.18 |
| Woodcock-Johnson: reading cluster | 0.27 | 0.46 | 0.49 | 0.00 | 0.87 | 0.17 |
| Letter/word identification | 0.28 | 0.46 | 0.48 | 0.00 | 0.88 | 0.18 |
| Reading comprehension | 0.27 | 0.32 | 0.30 | 0.04 | 0.88 | 0.16 |
| Woodcock-Johnson: math cluster | 0.25 | 0.42 | 0.34 | 0.07 | 0.46 | 0.22 |
| Applied problems (math, standard score) | 0.45 | 0.50 | 0.39 | 0.06 | 0.61 | 0.17 |
| WRAT-3: spelling (standard score) | 0.11 | 0.13 | 0.17 | 0.02 | 0.42 | 0.11 |
| Visual-motor integration | ||||||
| VMI (standard score) | 0.06 | 0.18 | 0.18 | 0.02 | 0.24 | 0.05 |
| VMI: other | 0.12 | 0.41 | 0.47 | 0.05 | 0.55 | 0.08 |
| Obsessive-compulsive behavior | ||||||
| CBC: No. of categories represented | 0.18 | 0.12 | 0.07 | 0.01 | 0.32 | 0.07 |
| CBC: interference with social | 0.03 | 0.46 | 0.60 | 0.18 | 0.73 | 0.36 |
| CBC: if interrupted–halts and resumes | 0.27 | 0.24 | 0.22 | 0.02 | 0.45 | 0.22 |
| CBC: if interrupted–other | 0.29 | 0.54 | 0.37 | 0.36 | 0.69 | 0.20 |
The coefficient of determination is the proportion of a sample variance of a response variable that is “explained” by the predictor variables when a linear regression is performed. The evaluation of the genes in explaining the variation in phenotypic measures was investigated by fitting a general linear model and specifying gene-expression (CT) values as the predictor variable and the behavioral or cognitive score as the response variable. Therefore, larger coefficient of determination values indicate a greater impact. The GCP5 gene generally produced smaller coefficient of determination scores and, thus, has less influence on the behavioral and academic parameters measured. WAIS-R indicates Wechsler Adult Intelligence Scale-Revised25; WRAT-3, Wide-Range Achievement Test, 3rd Edition.
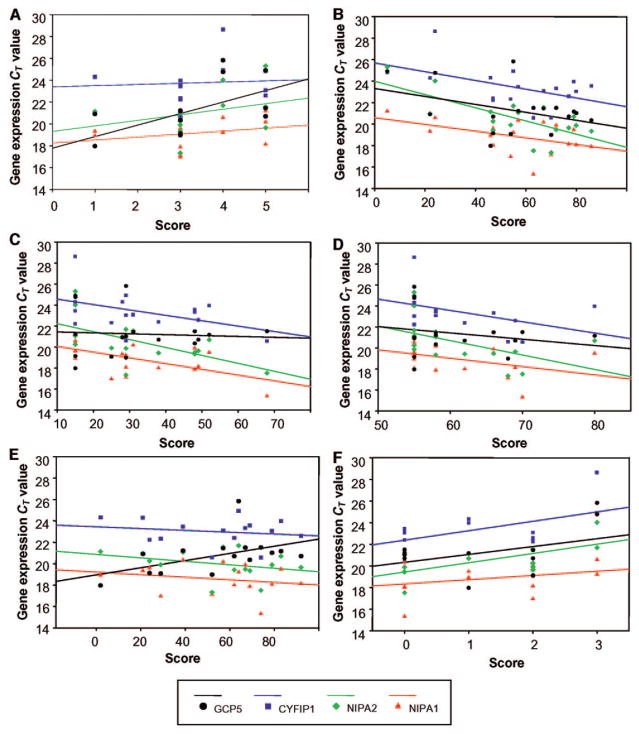
FIGURE 3.
Correlations of gene expression (based on CT values, which are inversely related to gene expression) for NIPA1, NIPA2, CYFIP1, and GCP5 and representative behavioral and cognitive parameters (A, SIB disruptive behavior frequency; B, Woodcock-Johnson math cluster; C, SIB broad independence score [standard score]; D, VMI [standard score]; E, reading comprehension; F, CBC if interrupted– other) between subjects with PWS with TI and TII deletions. Correlation values are provided in Table 2.
DISCUSSION
We quantified mRNA levels in lymphoblastoid cell cultures for the 4 genes (NIPA1, NIPA2, CYFIP1, and GCP5) between BP1 and BP2, which are haploinsufficient in subjects with PWS with TI deletions but not deleted in individuals with PWS having TII deletions. The mRNA of each of the 4 genes was significantly reduced in subjects with TI deletions compared with those with TII deletions (Table 1). The mRNA steady-state level can be affected by many cellular processes including transcription and turnover rate. We present here the relationship between mRNA quantity (ie, CT value) and behavioral and cognitive parameters. The quantity of mRNA of each 1 of the 4 genes was reduced by approximately fourfold to ~25% of that of the subjects with TII deletions. Because mRNA was reduced by >50% when the paternal allele was missing, the paternal allele may be more active than the maternal allele, but more studies are needed to confirm paternal bias.
We fit a linear-regression model to each parameter using the mean CT value (which is inversely related to quantity of mRNA) of each of the 4 genes as covariates. The coefficient of determination indicated that the quantity of mRNA of the 4 genes explained from 24% to 99% of the variation of the behavioral and academic parameters measured. By comparison, the coefficient of determination for deletion type alone explained from 5% to 50% of the variation in the assessed parameters. Thus, the covariation of quantity of mRNA explained the phenotypic variation more effectively than did deletion type (TI or TII). In addition, the variability in the coefficient of determination of the linear model for each gene suggests that the impact of the 4 genes varies with the phenotypic parameter. The expression of these genes may be impacting the parameters analyzed; not surprisingly, however, they certainly cannot be the only influence.
Although all 4 of the genes examined seemed to contribute to some degree to the parameters measured, NIPA2 seemed to have the greatest impact (Table 2), because a larger number of phenotypic parameters were noted with significant correlations with NIPA2 mRNA levels. Our data further suggest that NIPA1, NIPA2, and CYFIP1 may have a greater influence on the studied behavioral and cognitive parameters than does GCP5. Similarly, these 3 genes have been implicated in central nervous system development and/or function. NIPA1 has been associated with spastic paraplegia,17 whereas NIPA2 is conserved in vertebrates and widely expressed, including in the central nervous system. NIPA2 contains sequences with similarity to transmembrane domains, suggesting receptor or transporter function.5 Furthermore, The CYFIP1 protein is present in synaptosomal extracts and interacts with FMRP,29 the product of the FMR1 gene, which is responsible for fragile X syndrome. Understanding the influence of gene expression on behavioral and cognitive characteristics in humans is at an early stage of research development. Additional research is needed, including gene-expression studies in brain tissue from subjects with PWS, to identify the function of these genes and their interaction with gene networks to clarify the potential role they play in central nervous system development and function.
Acknowledgments
This research was partially supported by National Institute of Child Health and Human Development grants RO1 HD41672 and PO1 HD30329 and by the Hall Foundation of Kansas City.
We thank Steve Simon, PhD, for help with the statistical analysis.
Abbreviations
- PWS
Prader-Willi syndrome
- UPD
uniparental maternal disomy 15
- BP
breakpoint
- TI
type I
- TII
type II
- mRNA
messenger RNA
- CBC
Compulsive Behavior Checklist
- SIB
Scales of Independent Behavior
- VMI
Visual Motor Integrations
- RT
reverse transcription
- PCR
polymerase chain reaction
Footnotes
The authors have indicated they have no financial relationships relevant to this article to disclose.
References
- 1.Butler MG, Thompson T. Prader-Willi syndrome: clinical and genetic findings. Endocrinologist. 2000;10:35–165. doi: 10.1097/00019616-200010041-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassidy SB, Forsythe M, Heeger S, et al. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am J Med Genet. 1997;68:433–440. [PubMed] [Google Scholar]
- 3.Bittel D, Butler MG. Prader-Willi syndrome: clinical genetics, cytogenetics and molecular biology. Expert Rev Mol Med. 2005;7(14):1–20. doi: 10.1017/S1462399405009531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 5.Chai JH, Locke DP, Greally JM, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet. 2003;73:898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujana MA, Nadal M, Gratacos M, et al. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11–q13, 15q24, and 15q26. Genome Res. 2001;11:98–111. doi: 10.1101/gr.155601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler MG. Hypopigmentation: a common feature of Prader-Labhart-Willi syndrome. Am J Hum Genet. 1989;45:140–146. [PMC free article] [PubMed] [Google Scholar]
- 8.Butler MG. Prader-Willi syndrome: current understanding of cause and diagnosis. Am J Med Genet. 1990;35:319–332. doi: 10.1002/ajmg.1320350306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roof E, Stone W, MacLean W, Feurer ID, Thompson T, Butler MG. Intellectual characteristics of Prader-Willi syndrome: comparison of genetic subtypes. J Intellect Disabil Res. 2000;44:25–30. doi: 10.1046/j.1365-2788.2000.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassidy SB. Prader-Willi syndrome. J Med Genet. 1997;34:917–923. doi: 10.1136/jmg.34.11.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dykens EM, Cassidy SB, King BH. Maladaptive behavior differences in Prader-Willi syndrome due to paternal deletion versus maternal uniparental disomy. Am J Ment Retard. 1999;104:67–77. doi: 10.1352/0895-8017(1999)104<0067:MBDIPS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Gunay-Aygun M, Heeger S, Schwartz S, Cassidy SB. Delayed diagnosis in patients with Prader-Willi syndrome due to maternal uniparental disomy 15. Am J Med Genet. 1997;71:106–110. [PubMed] [Google Scholar]
- 13.Fox R, Yang GS, Feurer ID, Butler MG, Thompson T. Kinetic form discrimination in Prader-Willi syndrome. J Intellect Disabil Res. 2001;45:317–325. doi: 10.1046/j.1365-2788.2001.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113:565–573. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milner KM, Craig EE, Thompson RJ, et al. Prader-Willi syndrome: intellectual abilities and behavioural features by genetic subtype. J Child Psychol Psychiatry. 2005;46:1089–1096. doi: 10.1111/j.1469-7610.2005.01520.x. [DOI] [PubMed] [Google Scholar]
- 16.Varela MC, Kok F, Setian N, Kim CA, Koiffmann CP. Impact of molecular mechanisms, including deletion size, on Prader-Willi syndrome phenotype: study of 75 patients. Clin Genet. 2005;67:47–52. doi: 10.1111/j.1399-0004.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- 17.Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman WK, Price LH, Rasmussen SA, et al. The Yale-Brown Obsessive Compulsive Scale. II. Validity. Arch Gen Psychiatry. 1989;46:1012–1016. doi: 10.1001/archpsyc.1989.01810110054008. [DOI] [PubMed] [Google Scholar]
- 19.Goodman WK, Price LH, Rasmussen SA, et al. The Yale-Brown Obsessive Compulsive Scale. I. Development, use, and reliability. Arch Gen Psychiatry. 1989;46:1006–1011. doi: 10.1001/archpsyc.1989.01810110048007. [DOI] [PubMed] [Google Scholar]
- 20.Stein DJ, Keating J, Zar HJ, Hollander E. A survey of the phenomenology and pharmacotherapy of compulsive and impulsive-aggressive symptoms in Prader-Willi syndrome. J Neuropsychiatry Clin Neurosci. 1994;6:23–29. doi: 10.1176/jnp.6.1.23. [DOI] [PubMed] [Google Scholar]
- 21.Dykens EM, Leckman JF, Cassidy SB. Obsessions and compulsions in Prader-Willi syndrome. J Child Psychol Psychiatry. 1996;37:995–1002. doi: 10.1111/j.1469-7610.1996.tb01496.x. [DOI] [PubMed] [Google Scholar]
- 22.Gedye A. Issues involved in recognizing obsessive-compulsive disorder in developmentally disabled clients. Semin Clin Neuropsychiatry. 1996;1:142–147. doi: 10.1053/SCNP00100142. [DOI] [PubMed] [Google Scholar]
- 23.Reiss S. Test Manual for the Reiss Screen for Maladaptive Behavior. Orlando Park, IL: International Diagnostic Systems; 1988. [Google Scholar]
- 24.Bruininks RH, Woodcock RW, Weatherman RF, Hill BK. Scales of Independent Behavior (SIB) Allen, TX: DCM Teaching Resources; 1984. [Google Scholar]
- 25.Wechsler D. Wechsler Adult Intelligence Scale-Revised. New York, NY: The Psychological Corporation; 1981. [Google Scholar]
- 26.Wechsler D. Wechsler Intelligence Scale for Children-III. New York, NY: The Psychological Corporation; 1991. [Google Scholar]
- 27.Beery KE, Buktenica NA. Developmental Test of Visual-Motor Integration. Cleveland, OH: Modern Curriculum Press; 1997. 3rd revision. [Google Scholar]
- 28.Woodcock RW, Johnson MB. Woodcock-Johnson Tests of Achievment–Revised. Allen, TX: DLM Teaching Resources; 1990. [Google Scholar]
- 29.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]