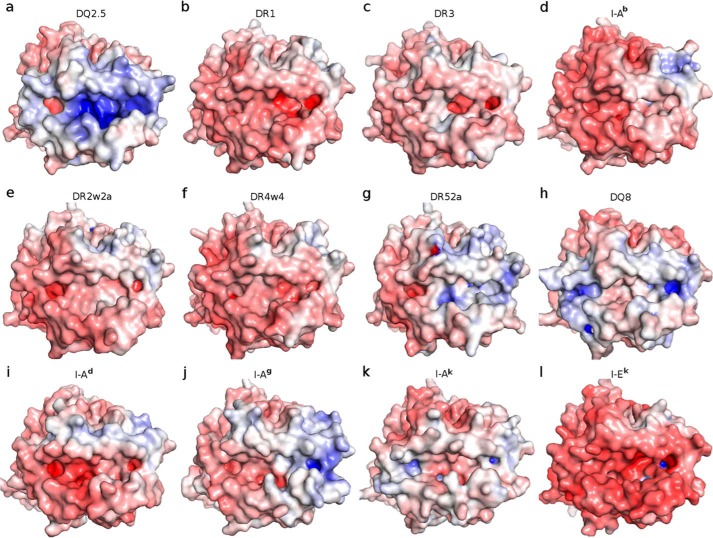
Figure 5.
Adaptive Poisson Boltzmann Solver-generated electrostatic surface of MHC class-II proteins at pH 7.0 (red, negative; blue, positive; white, neutral). The view is from the top, looking straight into the peptide-binding groove. a, DQ2.5·CLIP1 (Protein Data Bank code 5KSU); b, DR1·CLIP1 (Protein Data Bank code 3PDO); c, DR3·CLIP1 (Protein Data Bank code 1A6A); d, I-Ab·CLIP1 (Protein Data Bank code 1MUJ); e, DR2w2a·Epstein-Barr virus DNA polymerase peptide (Protein Data Bank code 1H15); f, DR4w4·human collagen II peptide (Protein Data Bank code 2SEB); g, DR52a-integrin β3 peptide (Protein Data Bank code 2Q6W); h, DQ8·deamidated gluten peptide (Protein Data Bank code 2NNA); i, I-Ad·influenza hemagglutinin peptide (Protein Data Bank code 2IAD); j, I-Ag·hen egg lysozyme(11–27) peptide (Protein Data Bank code 3MBE); k, I-Ak·conalbumin peptide (Protein Data Bank code 1D9K); l, I-Ek (moth cytochrome c) peptide (Protein Data Bank code 3QIU). MHC-bound peptides were not included in the Adaptive Poisson Boltzmann Solver electrostatics calculations.