Abstract
Estimation of intra- and interspecific interactions from time-series on species-rich communities is challenging due to the high number of potentially interacting species pairs. The previously proposed sparse interactions model overcomes this challenge by assuming that most species pairs do not interact. We propose an alternative model that does not assume that any of the interactions are necessarily zero, but summarizes the influences of individual species by a small number of community-level drivers. The community-level drivers are defined as linear combinations of species abundances, and they may thus represent e.g. the total abundance of all species or the relative proportions of different functional groups. We show with simulated and real data how our approach can be used to compare different hypotheses on community structure. In an empirical example using aquatic microorganisms, the community-level drivers model clearly outperformed the sparse interactions model in predicting independent validation data.
Keywords: community dynamics, density dependence, Gompertz model, interaction network, joint species distribution model, temporal analysis
1. Introduction
Biotic interactions are one of the principal drivers structuring species communities [1,2]. Individuals interact with members of their own species through density-dependent regulation [3], and with members of other species through e.g. interspecific competition, predation and facilitation [4–6]. Population dynamic models fitted to single species time-series data have demonstrated that population growth rate is density-dependent [7], due e.g. to increased mortality or decreased fecundity at times of high population density [8]. Multispecies population studies have shown that population fluctuations of interacting species can also influence population growth rates [9–11]. However, the contribution of biotic interactions in shaping complex and species-rich communities through time remains poorly explored in the ecological literature [12], partly due to the lack of effective statistical frameworks for analysing time-series data of large species communities.
On the one hand, applications of standard multivariate time-series models have enabled researchers to infer how intra- and interspecific interactions determine population dynamics only for small communities of a few interacting species [13–16]. This is because the number of all potential pairwise interactions among the species is vast for large communities and thus standard time-series models become difficult to estimate from limited data. On the other hand, a plethora of indices have been proposed to describe co-occurrence patterns among species for large communities [17–19], but such indices do not provide much insight into the underlying mechanisms driving community dynamics. Therefore, one of the key statistical challenges in ecology is to develop robust techniques that allow one to separate the directional and structural changes from natural temporal variation caused by intra- and interspecific interactions versus environmental stochasticity from time-series data on species-rich communities [13,20,21].
Multivariate autoregressive (MAR) models, also called vector autoregressive models (VAR), have become the most widely applied class of time-series models in community ecology [22,23]. Their use thus far, due to the curse of dimensionality, has however been restricted to small communities [24], to most common species only [13–16], or to a priori defined groups of species [20,22,25,26]. One solution that has been proposed to overcome the curse of dimensionality in MAR models is to constrain the estimation of the interaction matrix based on prior information about the existence and direction of interactions among specific pairs of species [22]. In the absence of such prior information, an alternative solution is to assume that most species do not interact, i.e. that a large proportion of the elements of the interaction matrix are zero, and use a variable selection procedure to identify the non-zero elements [13].
In this paper, we propose a new approach to estimating interaction matrices based on time-series data from species-rich communities. Our approach does not involve the assumption that any of the interactions are necessarily zero, but that the influences of the other species on the dynamics of a focal species can be summarized through a few community-level drivers. By community-level drivers we mean those linear combinations of species abundances that are most relevant in determining the future growth rates of all the species. Biologically, community-level drivers can for example represent the total abundance of all species (coefficients of linear combination equal for all species), the total biomass of the community (coefficients proportional to mass of each species), or different functional groups (coefficients non-zero only for a particular functional group). However, instead of determining a priori the contributions of the species to the community-level drivers (i.e. the coefficients of the linear combinations), we estimate them in a way that they best explain the data jointly for all species. To do so, we utilize recent developments in statistical literature on row–column interaction models [27].
Our approach is related to latent variable modelling, which has recently emerged in the ecological literature as a tool for estimating large co-occurrence matrices from snapshot data with joint species distribution models [28–32]. The explicit time-series model that we construct here can be seen as a more mechanistic alternative to a model in which the species would respond to temporally structured latent variables. While a latent variable model would necessarily lead to symmetric associations among the species (if species A influences species B positively, then species B necessarily influences species A positively), the explicit time-series model relaxes this assumption. In other words, while co-occurrence matrices are constrained to be symmetric and positive-definite, there is no such restriction for the interaction matrices, and thus the method presented here is technically related but not identical to latent variable approaches used for estimating co-occurrence matrices.
We compare the performance of the ‘community-level drivers' approach to previously published MAR approaches [13,22] using both simulated and real data. We first consider a set of simulated communities that differ in their size (i.e. number of species) and the underlying structure of the interaction matrix, and ask how well different approaches are able to (i) infer the interaction matrix and (ii) predict independent validation data. We then apply four alternative statistical models to a real time-series data on 100 species of aquatic microorganisms [33], to examine (iii) which of the statistical models performs best in predicting independent validation data, (iv) whether and how much accounting for interspecific interactions helps in predicting the validation data, and (v) what is the estimated structure of the interaction matrix for this community.
2. Methods
(a). Statistical modelling framework
We consider time-series data on species abundance that span over n + 1 time steps (e.g. years) and involve m species. We denote by yi,t the log-abundance of species i at time t, and by yt the vector for all species. We focus here on the standard first order multivariate autoregressive model MAR(1), defined by
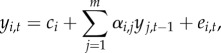 |
2.1 |
or equivalently in vector form by 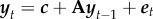 . The noise term is assumed to follow the multivariate normal distribution
. The noise term is assumed to follow the multivariate normal distribution 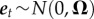 , independently among the time steps t. The intercept c (with elements ci) has the dimension m × 1, and the interaction matrix A (with elements αi,j) and the variance-covariance matrix Ω (with elements ωi,j) have the dimension m × m. Note that while the matrix Ω is symmetric, the matrix A may be asymmetric.
, independently among the time steps t. The intercept c (with elements ci) has the dimension m × 1, and the interaction matrix A (with elements αi,j) and the variance-covariance matrix Ω (with elements ωi,j) have the dimension m × m. Note that while the matrix Ω is symmetric, the matrix A may be asymmetric.
To connect the MAR(1) model to ecological literature, we note that equation (2.1) is mathematically equivalent to the widely applied Gompertz model [22,34], defined by
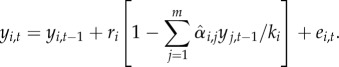 |
2.2 |
In the Gompertz model, yi,t is the log-abundance of species i at time t, ri is the growth rate and ki the carrying capacity of species i, and 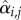 the influence of species j on species i. While we will follow here the parameterization of the MAR(1) model (equation (2.1)), its parameters can be mapped to those of the Gompertz model (equation (2.2)), and thus our results apply also to the latter model.
the influence of species j on species i. While we will follow here the parameterization of the MAR(1) model (equation (2.1)), its parameters can be mapped to those of the Gompertz model (equation (2.2)), and thus our results apply also to the latter model.
Two major limitations for ecological applications of the MAR(1) model are that it assumes the simplistic linear dependency on how the dynamics of a focal species are modified by other species, and that it assumes normally distributed residuals. Concerning the assumption of linearity, MAR(1) can be considered as an approximation to a more general class of nonlinear models [22]. With regard to the assumption of normality of residuals, we note that the model can be generalized to other data distributions by letting yi,t be the linear predictor within a generalized linear modelling framework. For example, Sebastián-González et al. [35] used the logit-link function to fit a generalized version of the MAR(1) model to presence–absence time-series data. Thus, while we develop our methods here in the context of the somewhat simplistic MAR(1) model, they can be applied also in a more general framework, e.g. allowing for the inclusion of sampling or observation error.
(i). Dimension reduction through community-level drivers
The parameterization of the MAR(1) model, and more generally any community-level time-series model, is challenging for large m, i.e. for species-rich communities. This is because the matrix A has m2 degrees of freedom and the symmetric matrix Ω has m(m + 1)/2 degrees of freedom. Thus, if not making some further structural assumptions, the parameterization of equation (2.1) requires very long time-series 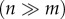 , which is unrealistic for real species-rich communities. Here we propose an alternative approach to the sparse interactions model [13] by not assuming that any of the interactions are necessarily zero, but that the interactions within the community are structured so that they can be described by a small number of community-level drivers. From the statistical point of view, our approach belongs to the class of row–column interaction models [27], which in turn are a special case of reduced rank vector models [36]. We model the community-level drivers as linear combinations of species occurrences,
, which is unrealistic for real species-rich communities. Here we propose an alternative approach to the sparse interactions model [13] by not assuming that any of the interactions are necessarily zero, but that the interactions within the community are structured so that they can be described by a small number of community-level drivers. From the statistical point of view, our approach belongs to the class of row–column interaction models [27], which in turn are a special case of reduced rank vector models [36]. We model the community-level drivers as linear combinations of species occurrences,
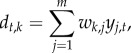 |
2.3 |
where dt,k is the community-level driver k (with 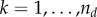 ) at time t, and wk,j is the contribution of species j to the driver k. Denoting by qi,k the influence of the community-level driver k on species i, the interaction terms of equation (2.1) can be written as
) at time t, and wk,j is the contribution of species j to the driver k. Denoting by qi,k the influence of the community-level driver k on species i, the interaction terms of equation (2.1) can be written as
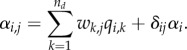 |
2.4 |
Here δij is Kronecker's delta (δii = 1 and δij = 0 for j ≠ i), and thus we have included separately a term for within species density dependence (αi) due to its obvious ecological importance. The advantage of equation (2.4) is that it greatly reduces be effective dimension of the interaction matrix, assuming that the number of community-level drivers is much smaller than the number of species: while in the original model the number of parameters in the interaction matrix A is m2, with equation (2.4) the matrix is constructed from m(2nd + 1) parameters. Consequently, the parameters of the model can be identified if the number of time steps is much greater than the amount of community-level drivers,  .
.
Similarly, the matrix Ω can be written with the help of latent factors (ηt,k) and factor loadings (λk,i) as
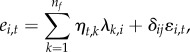 |
2.5 |
where 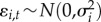 and nf is the number of latent factors. With this parameterization, it holds that
and nf is the number of latent factors. With this parameterization, it holds that 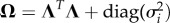 , where ʌ is the matrix of factor loadings
, where ʌ is the matrix of factor loadings 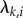 . As the parameterization of equation (2.5) has been discussed extensively in the context of joint species distribution modelling [32], we focus here mainly on the novel component of our work, i.e. equation (2.4).
. As the parameterization of equation (2.5) has been discussed extensively in the context of joint species distribution modelling [32], we focus here mainly on the novel component of our work, i.e. equation (2.4).
(ii). Alternative statistical frameworks
To evaluate the performance of the above described statistical model, we define a set of alternative models. We consider the following four models:
— Model 1: no interspecific interactions. In this model we assume that αi,j = 0 for i ≠ j.
— Model 2: full interactions. In this model, we estimate A as a full matrix without making any prior structural assumptions on it.
— Model 3: sparse interactions. Here we assume a priori that each off-diagonal element αi,j is non-zero with probability p whereas the diagonal elements αi,i are assumed to be non-zero.
— Model 4: community-level drivers. This is the model described in the previous section, and thus we model the interaction coefficients αi,j by the row–column interaction model of equation (2.4).
(iii). Model fitting
We parameterized the model in a Bayesian framework, implemented as an extension to HMSC-Matlab [32]. This implementation enables one not only to parameterize the model described above, but also to extend it to involve environmental covariates, species traits, phylogenetic relationships, as well as e.g. a spatially hierarchical or a spatially explicit study design. Further, in addition to normally distributed data, it includes as data models Bernoulli distribution (with probit link-function) for presence–absence data and Poisson and over-dispersed Poisson distributions (with log link-function) for count data. Concerning the prior distributions, as usual in factor analysis, we assumed that 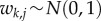 and
and 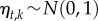 . We assumed a multiplicative gamma prior [37] for the influences of the community-level drivers (qi,k) and the latent factors (λk,i) on the species. In this model, the number of drivers nd is theoretically infinite, but their effective number is kept small due to increasing level of shrinkage applied to the influences of the community-level drivers (qi,k) as a function of the driver number. Due to computational reasons, the drivers that contribute a negligible proportion of variance are dropped from the model. In the sparse interactions model, we assumed that p = 0.1 as the default prior. For details on model fitting, see the electronic supplementary material.
. We assumed a multiplicative gamma prior [37] for the influences of the community-level drivers (qi,k) and the latent factors (λk,i) on the species. In this model, the number of drivers nd is theoretically infinite, but their effective number is kept small due to increasing level of shrinkage applied to the influences of the community-level drivers (qi,k) as a function of the driver number. Due to computational reasons, the drivers that contribute a negligible proportion of variance are dropped from the model. In the sparse interactions model, we assumed that p = 0.1 as the default prior. For details on model fitting, see the electronic supplementary material.
(b). Testing the performance of the approach with simulated data
To evaluate the performance of the statistical approach, we generated simulated data from the MAR(1) model. We conducted a full factorial design, in which we generated data using each of the above described four models, and then fitted to each dataset all the four models, resulting in 16 combinations of data-generating model and model used for inference. When generating data with the sparse interactions model, we set p = 0.1 and thus assumed that 90% of the interspecific interactions were zeros. When generating data with the community-level drivers model, we assumed that there were nd = 2 drivers. To test the influence of community size and length of time-series on the results, we assumed either a small (m = 5) or large community (m = 100), and either a short (n = 10) or long time-series (n = 100). We generated 10 replicates of each of these cases, thus resulting in 4 × 2×2 × 10 = 160 datasets and 4 × 160 = 640 models fitted to data. For details on data generation, see the electronic supplementary material.
We assessed the performances of the models both in terms of inference and predictive power. In terms of inference, we computed the correlation between the true and estimated (posterior mean) values of the interaction coefficients αi,j over all species pairs (i, j). In terms of predictive power, we predicted the posterior mean for yt, conditional on the true value of yt−1, for dynamics simulated for 100 additional time steps following the end of the time-series used for estimation. We computed the correlation between the predicted and observed values separately for each species, and then computed the average correlation over the species.
(c). An empirical case study
We analysed time-series data collected by Brannock et al. [38] on pelagic micro-eukaryote communities. The data were downloaded from the Dryad data repository [33]. The data originate from four sites that were bimonthly sampled during 2.5 years. In total, three sites were sampled 14 times and one site was sampled 10 times. The microorganisms were identified through high-throughput sequencing, the outcome of which was a matrix describing the sequence count for each OTU (operational taxonomical unit) for each site. Out of the 19 158 OTUs, we selected the 100 most common ones according to their prevalence (fraction of samples where the species was present), which varied among these 100 OTUs from 87% to 100%. We computed an abundance index by log(x + 1)-transforming the OTU count. We kept the last four time points for each site as validation data, and used the remaining data as training data. We fitted each of the Models 1–4 to the training data, including as covariates the season (fixed factor with the four levels of winter, spring, summer and fall), the site (random factor), the sample (random factor) and the log-transformed total number of sequences (continuous covariate). The last one was included to control for variation in sequencing depth, the total number of sequences per sample varying from 12 000 to 778 000. The random factors were implemented through a latent factor approach (equation (2.5)) following Ovaskainen et al. [32], and they model random variation in species occurrence and co-occurrence at these two levels. We evaluated the models' performances in terms of correlation between model prediction and validation data similarly to the evaluation of the simulated data. To test the sensitivity of the results to the prior assumptions related to the sparseness (Model 3) or effective dimensionality (Model 4) of the interaction matrix, we fitted the Models 3 and 4 also with alternative priors (see electronic supplementary material).
3. Results
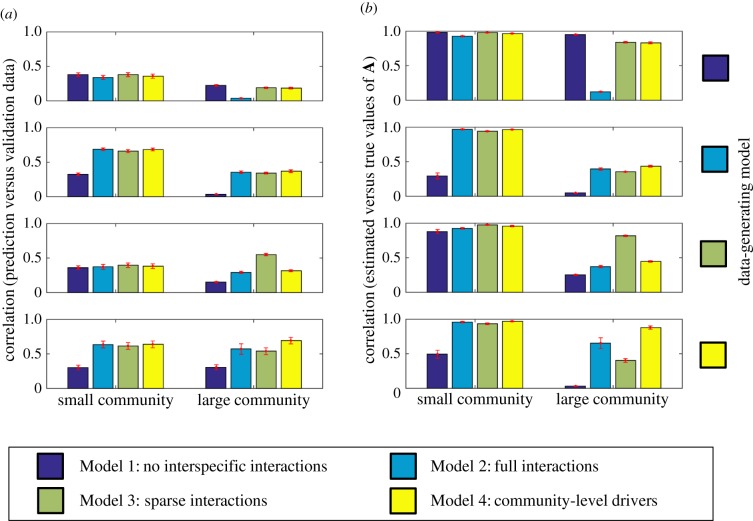
For a small community, long time-series data contains sufficient information for estimating the full A matrix. Consequently, for small communities, all models performed essentially equally well with simulated data both in terms of prediction (figure 1a) and inference (figure 1b). The exception is Model 1, which failed to perform well with data generated by the other models, simply because it assumes that the off-diagonal elements of A are zero. With large communities, which are the focus of this paper, the models deviated substantially from each other in their performance. As expected, the true model that was used to generate the data always performed at least equally well as the other models. Similarly to the case of the small communities, with large communities Model 1 performed well only if the underlying communities also lacked interspecific interactions. Among Models 2–4, Model 2 (full interactions) performed the worst: Models 3 and 4 performed equally well as Model 2 for data generated by Model 2, but Model 2 performed worse than Model 3 for data generated by Model 3, and it performed worse than Model 4 for data generated by Model 4. Neither Model 3 (sparse interactions) nor Model 4 (community-level drivers) was superior over the other one: Model 3 outperformed Model 4 with data generated by Model 3, whereas Model 4 outperformed Model 3 with data generated by Model 4.
Figure 1.
Comparison of the performance of the alternative statistical modelling frameworks based on simulated data. Panel (a) shows the correlation between model prediction and validation data (averaged over species), and panel (b) the correlation between elements of true and estimated (posterior mean) interaction matrices A. In both panels, the rows correspond to the data-generating models, the columns to small (m = 5) or large (m = 100) communities, and the colours to the models used for inference. The bars show the mean and the error bars ± two standard errors over the 10 replicates. The figure shows the results for a long time-series (n = 100), corresponding results for a short time-series (n = 10) being shown in the electronic supplementary material.
The results from simulated data demonstrate that the choice among the Models 3 and 4 is not a statistical but an ecological question: are real communities structured by sparse interactions or by interactions that can be captured by community-level drivers? While this question cannot be answered conclusively without the evaluation of a large array of case studies, the results from our empirical case study (with data illustrated in figure 2) give support for the community-level driver hypothesis: the correlations between model prediction and validation data were 0.37 (Model 1), 0.14 (Model 2), 0.32 (Model 3) and 0.46 (Model 4). Intriguingly, both Model 2 (full interactions) and Model 3 (sparse interactions) performed worse than Model 1 (no interspecific interactions). This is because of overfitting, which was the case for all models, and especially for Model 2: the correlation between model prediction and training data was 0.60 (Model 1), 0.98 (Model 2), 0.73 (Model 3), and 0.86 (Model 4). Repeating the analyses for alternative priors for Models 3 and 4 showed that the result of Model 4 outperforming Model 3 is robust to prior choice (electronic supplementary material). For additional evaluations of model fit, see electronic supplementary material.
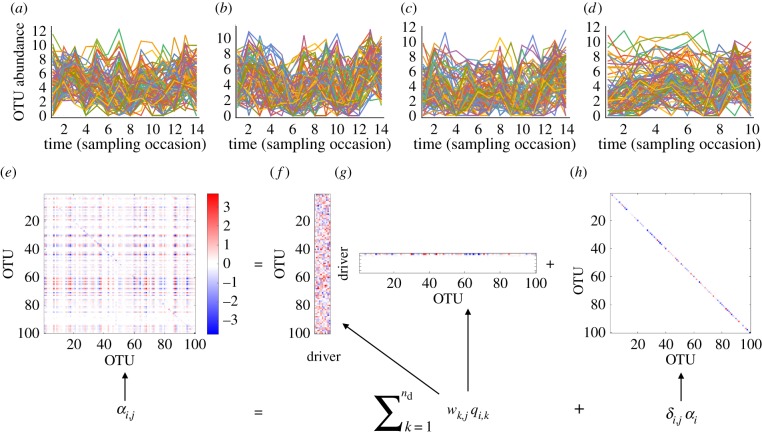
Figure 2.
Community-level drivers -model fitted to empirical data on aquatic micro-organisms. Panels (a–d) illustrate the log(x + 1)-transformed time-series data for the 100 most abundant OTUs on the four study sites. Panels (e–h) illustrate how the interaction matrix A is estimated through equation (2.4): the interaction matrix A shown in panel (e) is constructed through matrices representing the contributions of the species to the community-level drivers (wk,j, shown in panel f), the influences of the drivers to the species (qi,k, shown in panel g) and terms modelling within-species density dependence (αi, shown in panel h). While panel (e) shows the posterior mean estimate, the panels (f–h) show the posterior sample from the last Markov chain Monte Carlo round. This is because the terms wk,j and qi,k are not identifiable, as e.g. multiplying both of these by −1 leads to identical matrix A.
The contributions of the OTUs to each driver (wk,j) are illustrated in figure 2f, and the influences of the drivers to each species (qi,k) are illustrated in figure 2g. Effectively, the model identified only a single driver, as the influence of the first factor (top row in figure 2g) contributed 98.6% of the total influence of all 11 drivers that were included in the model. Thus, while Model 1 (full interactions) had 10 000 free parameters for the estimation of the matrix A, the community-level drivers model had essentially (counting the first driver only) only 300 parameters (100 parameters for each of wk,j, qi,k, αi). The terms modelling within-species density dependence αi (figure 2h) were clearly visible in the interaction matrix A, which had roughly equally many positive and negative off-diagonal elements (figure 2e). The matrix A was sparse in the sense that only a small fraction of the off-diagonal elements αi,j were estimated to be positive or negative with high statistical support: the fraction of elements that were positive with at least 95% posterior probability was 0.5%, and similarly the fraction of elements that were negative with at least 95% posterior probability was 0.2%.
4. Discussion
The ‘community-level drivers’ approach presented in this paper provides a new statistical framework for using time-series data on large communities to identify biotic and environmental drivers structuring communities. The method introduced here enables ecologists to efficiently estimate interaction matrices for species-rich communities, and thus to get a more accurate picture of interspecific interaction networks than so far has been possible. Large-scale and long-term time-series community data originating from environmental barcoding techniques are becoming increasingly available [39,40], and thus there is an increasing demand for robust statistical tools for analysing such large data. When combined with earlier developments in joint species distribution modelling [30–32], the statistical methods developed and implemented here enable analyses of such data in a way that integrates information on community-level dynamics with environmental covariates, species traits, phylogenetic relationships and spatial structure (e.g. spatially hierarchical or spatially explicit study designs). Estimated parameters may subsequently be used to evaluate the relative importance of intra- and interspecific interactions, as well as the stability of a community [22]. Moreover, since the method presented is a model-based approach, it can be used to predict community dynamics under environmental change, which is a key priority in conservation biology [41].
The community-level driver and the sparse interactions approaches represent two different ways to deal with the curse of the dimensionality problem encountered when estimating large interaction matrices. These two approaches may also be considered as alternative hypotheses about the structures of ecological interaction networks. Results from simulations performed here indicate that both approaches are able to predict community dynamics better than the full interaction model, but we acknowledge that for a comprehensive evaluation, more simulations under varying sets of assumptions and parameters as well as tests on empirical data are needed. The methods presented here allow one to test among these competing hypotheses, by fitting the competing models to data and comparing predictive performances of the models or applying other model selection approaches. The case study considered here gave support for the interactions between aquatic microorganisms being structured more closely according to ‘the community-level drivers hypothesis’ rather than ‘the sparse interactions hypothesis’. An important challenge for community ecological research is to disentangle these, and possibly other hypotheses, for a broad range of taxa and environmental settings. The community driver approach may also be extended to test further hypotheses, such as analysing whether the effects among trophically similar species on total abundance are equal, as proposed by the neutral theory [42]. This can be done by imposing constraints on the interaction matrix (equations (2.3–2.4)). Furthermore, our approach also provides tools for validating the critical assumptions regarding parametric species abundance models used to analyse temporal variation in community structure [43].
In the empirical case study involving aquatic microorganisms, we found strong statistical support for a positive or negative interaction only for very few species pairs. This finding is in line with theoretical and empirical studies showing that compared to the effects of the environment and intraspecific interactions, the contributions of interspecific interactions in structuring ecological communities are weak [13,44,45]. In spite of this, accounting for interspecific interactions greatly improved the predictive performance of the model, the correlation between model prediction and validation data increasing from 0.37 (Model 1) to 0.46 (Model 4). Curiously, even if the sparse interaction model is designed for a case where most interactions are zero, this approach led to even worse predictive power than Model 1, which sets all interactions to zero. Together, these results illustrate that, in the case of the community-level drivers model, the joint posterior distribution of the interaction matrix involves more information than what might be inferred from the marginal distributions of the interaction coefficients for each species pair. This is perhaps not surprising, as the model was not designed to capture interactions among specific species pairs, but structural properties of the interaction matrix. This is consistent with prior information on the importance of trophic interactions in the taxa relevant for the empirical study [46], e.g. large zooplankton being predators of small zooplankton and protists, and small zooplankton being grazers of phytoplankton.
Interaction networks might be structured by species traits [14,47], and thus modelling the species contributions to community-level drivers as well as the species responses to them as a function of traits is an important challenge for the future. Further aspects not considered here but potentially important in determining population dynamics include e.g. demographic stochasticity, migration and age structure. Incorporating these factors into the modelling framework in addition to mechanisms already included would further improve our mechanistic understanding of community dynamics.
Supplementary Material
Acknowledgements
We thank Mark Vellend and two anonymous reviewers for excellent comments on earlier versions of the manuscript.
Data accessibility
The empirical data used in this study were obtained from the Dryad public data repository. The HMSC 2.1 MATLAB software, the user manual and the scripts for replicating the analyses presented here are found from https://www.helsinki.fi/en/researchgroups/metapopulation-research-centre/hmsc21.
Authors' contributions
O.O. and D.D. conceived the statistical method, and O.O. and G.T. implemented it to HMSC-Matlab. O.O. constructed the numerical examples of the paper. N.A. and O.O. wrote the first draft of the paper, and all authors contributed to the writing of the final version.
Competing interests
We declare we have no competing interests.
Funding
The research was funded by the Academy of Finland (CoE grant no. 284601 to O.O.), the Research Council of Norway (SFF-III grant no. 223257) and the LUOVA graduate school of the University of Helsinki (PhD grant for G.T.).
References
- 1.Götzenberger L, et al. 2012. Ecological assembly rules in plant communities—approaches, patterns and prospects. Biol. Rev. 87, 111–127. ( 10.1111/j.1469-185X.2011.00187.x) [DOI] [PubMed] [Google Scholar]
- 2.Vellend M. 2010. Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206. ( 10.1086/652373) [DOI] [PubMed] [Google Scholar]
- 3.Hassell MP. 1975. Density-dependence in single-species populations. J. Anim. Ecol. 44, 283–295. ( 10.2307/3863) [DOI] [Google Scholar]
- 4.Holt RD, Polis GA. 1997. A theoretical framework for intraguild predation. Am. Nat. 149, 745–764. ( 10.1086/286018) [DOI] [Google Scholar]
- 5.Gallagher ED, Jumars PA, Trueblood DD. 1983. Facilitation of soft-bottom benthic succession by tube builders. Ecology 64, 1200–1216. ( 10.2307/1937829) [DOI] [Google Scholar]
- 6.Tilman D. 1994. Competition and biodiversity in spatially structured habitats. Ecology 75, 2–16. ( 10.2307/1939377) [DOI] [Google Scholar]
- 7.Brook BW, Bradshaw CJA. 2006. Strength of evidence for density dependence in abundance time series of 1198 species. Ecology 87, 1445–1451. ( 10.1890/0012-9658%282006%2987%5B1445%3ASOEFDD%5D2.0.CO%3B2) [DOI] [PubMed] [Google Scholar]
- 8.Sæther B, et al. 2016. Demographic routes to variability and regulation in bird populations. Nat. Commun. 7, 12001 ( 10.1038/ncomms12001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt RD. 1977. Predation, apparent competition, and the structure of prey communities. Theor. Popul. Biol. 12, 197–229. ( 10.1016/0040-5809(77)90042-9) [DOI] [PubMed] [Google Scholar]
- 10.Ives AR. 1995. Predicting the response of populations to environmental change. Ecology 76, 926–941. ( 10.2307/1939357) [DOI] [Google Scholar]
- 11.Klug JL, Fischer JM, Ives AR, Dennis B. 2000. Compensatory dynamics in planktonic community responses to pH perturbations. Ecology 81, 387–398. ( 10.2307/177435) [DOI] [Google Scholar]
- 12.Novak M, Yeakel JD, Noble AE, Doak DF, Emmerson M, Estes JA, Jacob U, Tinker MT, Wootton JT. 2016. Characterizing species interactions to understand press perturbations: what is the community matrix? Annu. Rev. Ecol. Evol. Syst. 47, 409–432. ( 10.1146/annurev-ecolsys-032416-010215) [DOI] [Google Scholar]
- 13.Mutshinda CM, O'Hara RB, Woiwod IP. 2009. What drives community dynamics? Proc. R. Soc. B 276, 2923–2929. ( 10.1098/rspb.2009.0523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almaraz P, Oro D. 2011. Size-mediated non-trophic interactions and stochastic predation drive assembly and dynamics in a seabird community. Ecology 92, 1948–1958. ( 10.2307/23034828) [DOI] [PubMed] [Google Scholar]
- 15.Durant JM, Krasnov YV, Nikolaeva NG, Stenseth NC. 2012. Within and between species competition in a seabird community: statistical exploration and modeling of time-series data. Oecologia 169, 685–694. ( 10.1007/s00442-011-2226-3) [DOI] [PubMed] [Google Scholar]
- 16.Porzig EL, Seavy NE, Eadie JM, Humple DL, Geupel GR, Gardali T. 2016. Interspecific interactions, population variation, and environmental forcing in the context of the community. Ecosphere 7, e01349 ( 10.1002/ecs2.1349) [DOI] [Google Scholar]
- 17.Magurran AE, McGill BJ. 2011. Biological diversity, 1st edn Oxford, UK: Oxford University Press. [Google Scholar]
- 18.Legendre P, Gauthier O. 2014. Statistical methods for temporal and space–time analysis of community composition data. Proc. R. Soc. B 281, 20132728 ( 10.1098/rspb.2013.2728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kara EL, Hanson PC, Hu YH, Winslow L, McMahon KD. 2013. A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. ISME J. 7, 680–684. ( 10.1038/ismej.2012.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martorell C, Freckleton RP. 2014. Testing the roles of competition, facilitation and stochasticity on community structure in a species-rich assemblage. J. Ecol. 102, 74–85. ( 10.1111/1365-2745.12173) [DOI] [Google Scholar]
- 21.Loreau M, de Mazancourt C. 2013. Biodiversity and ecosystem stability: a synthesis of underlying mechanisms. Ecol. Lett. 16, 106–115. ( 10.1111/ele.12073) [DOI] [PubMed] [Google Scholar]
- 22.Ives AR, Dennis B, Cottingham KL, Carpenter SR. 2003. Estimating community stability and ecological interactions from time-series data. Ecol. Monogr. 73, 301–330. ( 10.1890/0012-9615%282003%29073%5B0301%3AECSAEI%5D2.0.CO%3B2) [DOI] [Google Scholar]
- 23.Hampton SE, Holmes EE, Scheef LP, Scheuerell MD, Katz SL, Pendleton DE, Ward EJ. 2013. Quantifying effects of abiotic and biotic drivers on community dynamics with multivariate autoregressive (MAR) models. Ecology 94, 2663–2669. ( 10.1890/13-0996.1) [DOI] [PubMed] [Google Scholar]
- 24.Vik JO, Brinch CN, Boutin S, Stenseth NC. 2008. Interlinking hare and lynx dynamics using a century's worth of annual data. Popul. Ecol. 50, 267–274. ( 10.1007/s10144-008-0088-2) [DOI] [Google Scholar]
- 25.Hampton SE, Schindler DE. 2006. Empirical evaluation of observation scale effects in community time series. Oikos 113, 424–439. ( 10.1111/j.2006.0030-1299.14643.x) [DOI] [Google Scholar]
- 26.Gross K, Edmunds PJ. 2015. Stability of Caribbean coral communities quantified by long-term monitoring and autoregression models. Ecology 96, 1812–1822. ( 10.1890/14-0941.1) [DOI] [PubMed] [Google Scholar]
- 27.Yee TW, Hadi AF. 2014. Row–column interaction models, with an R implementation. Comput. Stat. 29, 1427–1445. ( 10.1007/s00180-014-0499-9) [DOI] [Google Scholar]
- 28.Ovaskainen O, Abrego N, Halme P, Dunson D. 2016. Using latent variable models to identify large networks of species-to-species associations at different spatial scales. Methods Ecol. Evol. 7, 549–555. ( 10.1111/2041-210X.12501) [DOI] [Google Scholar]
- 29.Thorson JT, Scheuerell MD, Shelton AO, See KE, Skaug HJ, Kristensen K. 2015. Spatial factor analysis: a new tool for estimating joint species distributions and correlations in species range. Methods Ecol. Evol. 6, 627–637. ( 10.1111/2041-210X.12359) [DOI] [Google Scholar]
- 30.Thorson JT, Ianelli JN, Larsen EA, Ries L, Scheuerell MD, Szuwalski C, Zipkin EF. 2016. Joint dynamic species distribution models: a tool for community ordination and spatio-temporal monitoring. Glob. Ecol. Biogeogr. 25, 1144–1158. ( 10.1111/geb.12464) [DOI] [Google Scholar]
- 31.Warton DI, Blanchet FG, O'Hara RB, Ovaskainen O, Taskinen S, Walker SC, Hui FKC. 2015. So many variables: joint modeling in community ecology. Trends Ecol. Evol. 30, 766–779. ( 10.1016/j.tree.2015.09.007) [DOI] [PubMed] [Google Scholar]
- 32.Ovaskainen O, Tikhonov G, Norberg A, Blanchet FG, Duan L, Dunson D, Roslin T, Abrego N. 2017. How to make more out of community data? A conceptual framework and its implementation as models and software. Ecol. Lett. 20, 561–576. ( 10.1111/ele.12757) [DOI] [PubMed] [Google Scholar]
- 33.Brannock PM, Ortmann AC, Moss AG, Halanych KM. 2016. Data from: Metabarcoding reveals environmental factors influencing spatio-temporal variation in pelagic micro-eukaryotes. Dryad Digital Repository. ( 10.5061/dryad.442dv) [DOI] [PubMed] [Google Scholar]
- 34.Royama T. 1992. Analytical population dynamics, 1st edn London, UK: Chapman & Hall. [Google Scholar]
- 35.Sebastián-González E, Sánchez-Zapata JA, Botella F, Ovaskainen O. 2010. Testing the heterospecific attraction hypothesis with time-series data on species co-occurrence. Proc. R. Soc. B 277, 2983–2990. ( 10.1098/rspb.2010.0244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yee TW, Hastie TJ. 2003. Reduced-rank vector generalized linear models. Stat. Model. 3, 15–41. ( 10.1191/1471082X03st045oa) [DOI] [Google Scholar]
- 37.Bhattacharya A, Dunson DB. 2011. Sparse Bayesian infinite factor models. Biometrika 98, 291–306. ( 10.1093/biomet/asr013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brannock PM, Ortmann AC, Moss AG, Halanych KM. 2016. Metabarcoding reveals environmental factors influencing spatio-temporal variation in pelagic micro-eukaryotes. Mol. Ecol. 25, 3593–3604. ( 10.1111/mec.13709) [DOI] [PubMed] [Google Scholar]
- 39.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. 2012. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. ( 10.1111/j.1365-294X.2012.05470.x) [DOI] [PubMed] [Google Scholar]
- 40.Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M. 2014. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367. ( 10.1016/j.tree.2014.04.003) [DOI] [PubMed] [Google Scholar]
- 41.Loreau M, et al. 2001. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804–808. ( 10.1126/science.1064088) [DOI] [PubMed] [Google Scholar]
- 42.Hubbell SP. 2001. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press. [Google Scholar]
- 43.Sæther B-E, Engen S, Grøtan V. 2013. Species diversity and community similarity in fluctuating environments: parametric approaches using species abundance distributions. J. Anim. Ecol. 82, 721–738. ( 10.1111/1365-2656.12068) [DOI] [PubMed] [Google Scholar]
- 44.Kokkoris GD, Troumbis AY, Lawton JH. 1999. Patterns of species interaction strength in assembled theoretical competition communities. Ecol. Lett. 2, 70–74. ( 10.1046/j.1461-0248.1999.22058.x) [DOI] [Google Scholar]
- 45.Mutshinda CM, O'Hara RB, Woiwod IP. 2011. A multispecies perspective on ecological impacts of climatic forcing. J. Anim. Ecol. 80, 101–107. ( 10.1111/j.1365-2656.2010.01743.x) [DOI] [PubMed] [Google Scholar]
- 46.D'Alelio D, Libralato S, Wyatt T, Ribera d'Alcalà M. 2016. Ecological-network models link diversity, structure and function in the plankton food-web. Sci. Rep. 6, 621806 ( 10.1038/srep21806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santamaría L, Rodríguez-Gironés MA. 2007. Linkage rules for plant–pollinator networks: trait complementarity or exploitation barriers? PLoS Biol. 5, e31 ( 10.1371/journal.pbio.0050031) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The empirical data used in this study were obtained from the Dryad public data repository. The HMSC 2.1 MATLAB software, the user manual and the scripts for replicating the analyses presented here are found from https://www.helsinki.fi/en/researchgroups/metapopulation-research-centre/hmsc21.