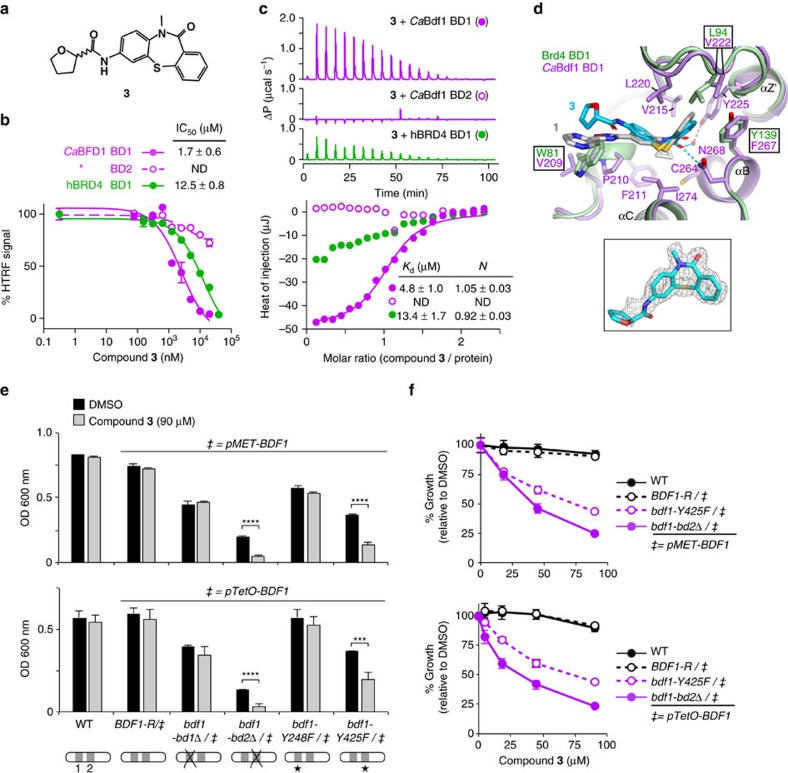
Figure 8. A CaBdf1 BD1 inhibitor phenocopies the effect of BD1 inactivation on C. albicans growth.
(a) Chemical structure of compound 3. (b) HTRF assay showing that 3 inhibits CaBdf1 BD1 preferentially over CaBdf1 BD2 and with modest selectivity over Brd4 BD1. (c) Representative ITC experiments showing the binding of 3 towards the indicated BD. The values indicated for Kd and N represent the mean and s.d. from three independent experiments. See also Supplementary Table 1. (d) Crystal structure of CaBdf1 BD1 (violet) bound to 3 (S enantiomer) and alignment with CaBdf1 BD1 bound to 1 and with human Brd4 BD1 (green). Side chains are shown for CaBdf1 residues in contact with 3 and for the corresponding Brd4 residues if divergent from CaBdf1. Inset, simulated-annealing omit Fo-Fc density for compound 3 contoured at 3σ. (e) Compound 3 compromises the in vitro growth of C. albicans when CaBdf1 BD2 is inactivated by deletion or point mutation. Strains were grown in the presence of methionine/cysteine or Dox to repress expression of the WT allele from the pMET (top) or pTetO (bottom) promoter, respectively. Data represent mean and s.d. values from three independent experiments. ***P≤0.01; ****P≤10−3. P values were determined in a two-sided Welch t-test. (f) Dose–response experiments showing the effect of 3 on Bdf1 mutant strains. Met/Cys or doxycyline were added to repress expression from the pMET (top) or pTetO (bottom) promoter, respectively. Data represent mean and s.d. values from three independent experiments.