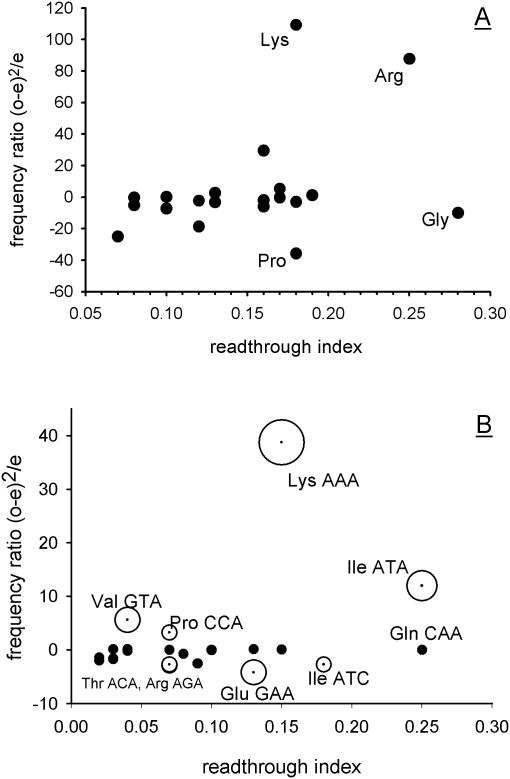
Figure 2.
Codon bias at the −1 and −2 codon positions preceding the stop codon is not predictive of effects on stop codon readthrough. Codon usage at the −1 and −2 codon positions preceding the stop codon was recorded for all S.cerevisiae ORFs. These observed frequencies (o) were compared with expected frequencies (e) derived from a codon usage table for all yeast ORFs. A frequency ratio [(o − e)2/e] was calculated for each of the codons at the −1 position. For the −2 position, the encoded amino acid frequency was recorded (the primary readthrough determinant at this position (15). The frequency ratio was assigned a polarity depending on whether the codon or encoded amino acid was either over-represented (positive polarity) or under-represented (negative polarity) relative to the expected frequency. (A) The −2 position codons were surveyed for all yeast genes, and the polar chi-squared values for each amino acid were plotted against the known effect of amino acid at this position on stop codon readthrough (15). (B) The process was repeated for −1 position codons, surveyed in a sub-set of genes containing other poor −2 position and stop codon context elements (see text for details). The significance level of under- or over-abundance of −1 position codons is indicated by the three sizes of bubbles used [0.001% (largest), 0.01% and 0.1% (smallest)].