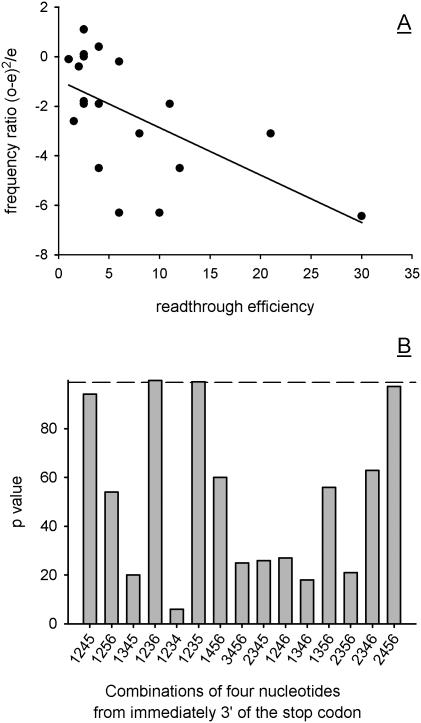
Figure 4.
Identifying key nucleotide components of the 3′ stop codon context. Fifteen different tetramer combinations of nucleotide positions (sub-contexts) from within the six nucleotide positions downstream of the stop codon were tested for an inverse correlation between tetramer sequence usage and known effects on stop codon readthrough. A set of 19 known 3′ contexts known to confer leakiness on the upstream stop codon was used to calibrate the predictive quality of each of the 15 sub-contexts. The frequency of these reference tetrameric sequences in control (3′-UTR) and stop codon flanking populations was compared, producing expected and observed frequencies, respectively. These were used to calculate frequency ratios [(o − e)2/e], which were plotted against known readthrough levels for the 22 reference 3′ contexts. The +1235 subcontext correlation between abundance in the genome and stop codon readthrough efficiency is shown (A). Linear regression analysis was carried out on this type of correlation plot for all 15 sub-context types. The regression p value obtained in each case was plotted in histogram form (B). For reference, the dotted line shows the 99% significance cut-off used.