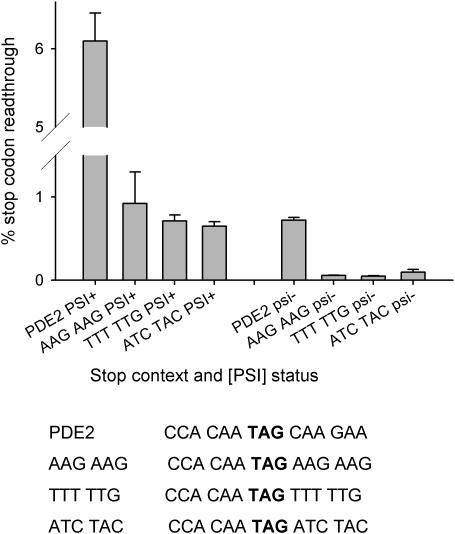
Figure 5.
Testing potential readthrough contexts identified through analysis of genome-wide 3′ contexts. Sequences of 6 nt identified in the 3′ context genome screen which represent potential poor 3′ stop codon contexts were tested for their ability to direct stop codon readthrough. The nucleotide hexamers AAGAAG and TTTTTG were used to replace the corresponding 3′ context element of the PDE2 stop codon environment placed in the pAC98 readthrough test system (Materials and Methods). The PDE2 3′ context was used as a positive control, and an over-represented 3′ sequence (ATCTAC) as a negative control. Levels of stop codon readthrough were determined in both [PSI+] and [psi−] derivatives of yeast strain 76D694. The mean of three readthrough determinations is shown (bars represent the SD, n = 3).