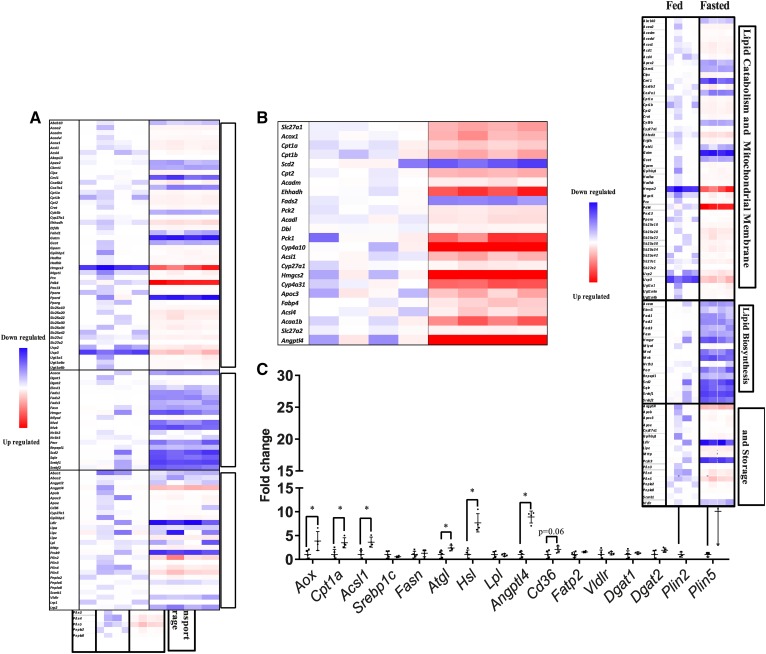
Fig. 2.
The kidney in the fasted state upregulates lipid oxidation genes and downregulates lipid synthesis genes. RNA sequencing of fed and fasted male kidneys. Data presented as fold change normalized to the fed group. DEGs were identified with Bioconductor package DEseq2 (3.2) and Benjamini and Hochberg’s method was used to control for false discovery rate. A: Canonical genes in pathways related to lipid metabolism and mitochondrial membranes, lipid biosynthesis and lipid transport, and lipolysis. B: Canonical genes in the PPAR pathway. C: Quantitative PCR of selected lipid metabolism genes, normalized to ribosomal 18s. Experiments were performed in male mice, N = 3–5. *P < 0.05. Results are presented as mean ± SD. Results for quantitative PCR were compared by unpaired Student’s t-test.