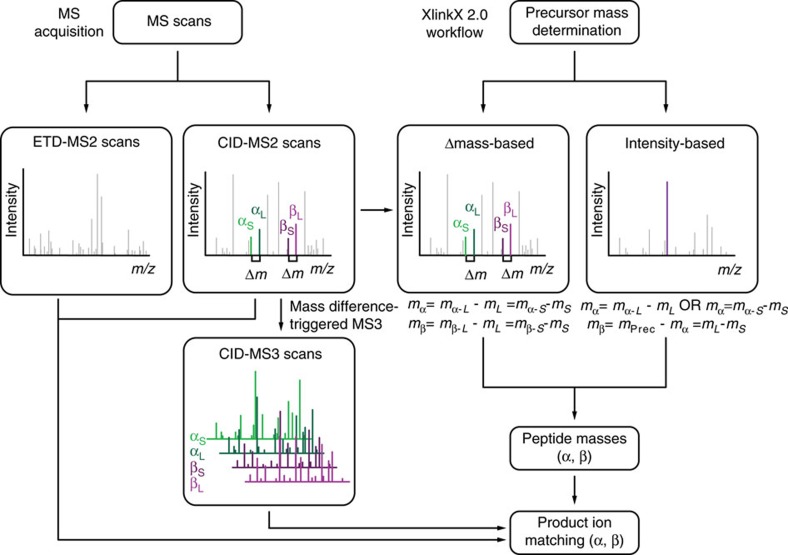
Figure 1. The multi-dimensional XL-MS data acquisition and analysis strategy.
Each MS1 precursor ion is subjected to sequential CID–MS2 and ETD–MS2 fragmentation. Data-dependent MS3 scans are performed if a unique mass difference (Δm) is found in the CID–MS2 scans. In XlinkX v2.0 data analysis, CID–MS2 scans are used to calculate the potential precursor mass of each linked peptide, using Δm-based, intensity-based or both strategies (see Methods). Both CID–MS2 and ETD–MS2 scans, as well as MS3 scans, are subjected to product ion matching to sequence the two peptide constituents of a cross-link. The four signature fragment ions with a unique mass difference (Δm), resulting from CID-induced cross-linker cleavage are shown in colours.