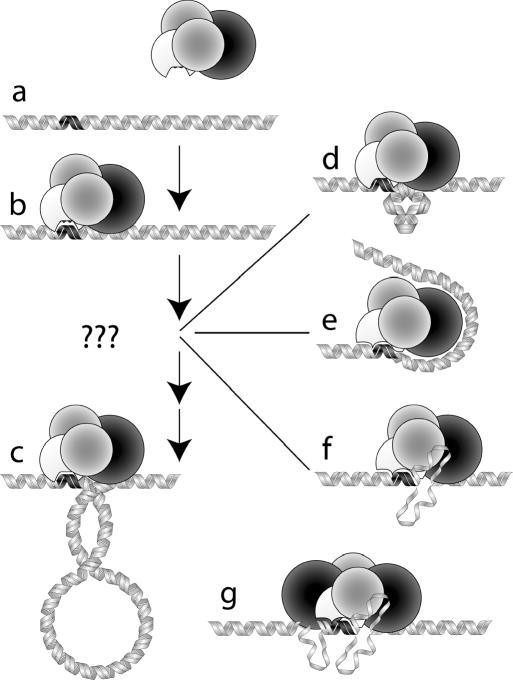
Figure 1.
The DNA translocation process for the R1-complex. (a) The black region on the DNA represents the DNA binding (recognition) site of the enzyme, which is represented by the four globular subunits of the R1-complex: HsdS (white), HsdM (grey) and HsdR (black). (b) HsdS is bound to the DNA at the recognition site and HsdR begins to contact adjacent DNA sequences. (c) The motor translocates adjacent DNA through the motor/DNA complex, which remains tightly bound to the recognition sequence. Translocation produces an expanding loop of negatively supercoiled DNA. In the early stages of translocation, a very small loop would require a significant energetic penalty and is thus unlikely. (d) The inherent stiffness of DNA prevents bending over short distances and a bulge of the type shown would require a large amount of energy from protein–DNA interactions. (e) Wrapping of DNA around the HsdR subunit in an attempt to overcome the problem associated with bending the relatively stiff DNA. (f) Alternatively, unwinding of DNA can be used to overcome the problems associated with the persistence length of DNA, resulting in an extrusion or a bubble of ssDNA. (g) R2 initial complexes exhibit a much larger, more intricate structure that is more difficult to analyse with AFM imaging.