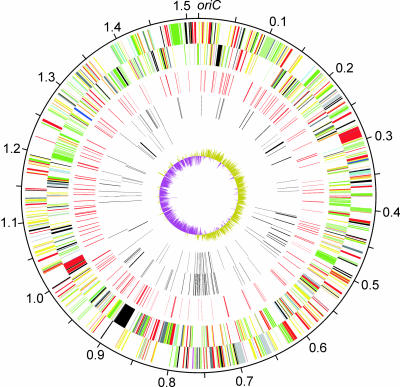
Fig. 1.
Circular representation of the genome of E. ruminantium (Welgevonden type strain). The outermost circle indicates the scale in megabases. The remaining concentric circles are described from outside to inside. The first and second circles represent the predicted coding sequences on the plus and minus strands, respectively, and are color-coded by function: dark blue, stable RNAs; black, chaperones and transporters; dark gray, energy metabolism; red, information transfer; yellow, central or intermediary metabolism; dark green, membrane proteins; cyan, degradation of large molecules; purple, degradation of small molecules; pale blue, regulators; orange, conserved hypothetical proteins; pink, phage and insertion sequence elements; brown, pseudogenes; pale green, unknown; light gray, miscellaneous. The third circle represents tandem repeats (shown in red). The fourth and fifth circles represent dispersed repeats (direct and inverted repeats, respectively), which are colored in black. The sixth circle represents G+C skew, with values greater than zero in olive and less than zero in magenta. A full-sized version of this figure is available as Fig. 4, which is published as supporting information on the PNAS web site.