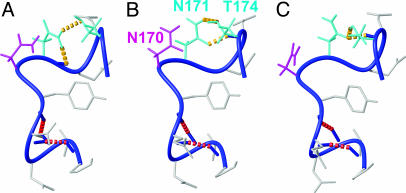
Fig. 3.
Local hydrogen-bonding polymorphisms in the loop comprising residues 165–175 in mPrP[S170N,N174T]. Three of the 20 dyana conformers used to represent the NMR structure (Table 1) are displayed. The backbone is a dark blue spline function through the Cα atoms, and side chains are gray, except for the following: The side chain (including hydrogen atoms) of N170 is shown in magenta, and the side chains of the hydrogen-bonded residues N171 and T174 are in cyan. The following hydrogen bonds in the three conformers are indicated with dashed yellow lines: Asn-171 Hδ1–Asn-171 O′ and Asn-171 Hδ2–Thr-174 Oγ (A), Asn-171 Hδ2–Thr-174 Oγ and Thr-174 HN–Asn-171 Oδ (B), and Thr-174 Hγ–Asn-171 Oδ and Thr-174 HN–Asn-171 Oδ (C). Two additional hydrogen bonds from Pro-165 O′ to Gln-168 HN and from Val-166 O′ to Tyr-169 HN (red broken lines), which define the 310-helical turn, are present in all 3 conformers [and in 19 of the 20 conformers used to represent the NMR structure (Table 1)].