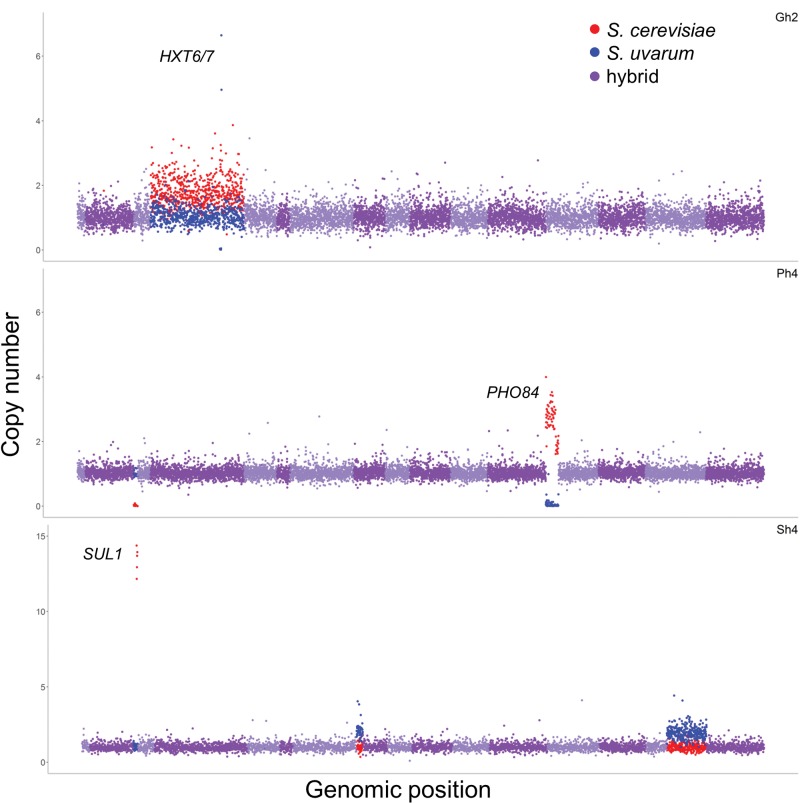
Fig. 1.
Evolved hybrids exhibit changes in copy number and loss of heterozygosity. Copy number variants are displayed for selected evolved hybrid clones from three nutrient-limited conditions: Gh2, glucose; Ph4, phosphate; and Sh4, sulfate. See additional figures in supplementary fig. S3, Supplementary Material online. Hybrid copy number, determined by normalized sequencing read depth per open reading frame (ORF), is plotted across the genome according to S. cerevisiae ORF coordinates to account for three reciprocal translocations between S. cerevisiae and S. uvarum. Chromosomes are plotted in alternating light and dark purple, red indicates a S. cerevisiae copy number variant, and blue indicates a S. uvarum copy number variant. Gh2 has a whole chromosome amplification of S. cerevisiae chrIV, a small segmental deletion of S. uvarum chrIV (non-copy neutral loss of heterozygosity), and an amplification of S. uvarum HXT6/7. Ph4 has a small segmental deletion of S. cerevisiae chrIII (non-copy neutral loss of heterozygosity) and an amplification of S. cerevisiae chrXIII with corresponding deletion of S. uvarum chrXIII (copy neutral loss of heterozygosity). Sh4 has an amplification of S. cerevisiae SUL1 and a whole chromosome amplification of S. uvarum chrVIII (note, there is a reciprocal translocation between chrVIII and chrXV in S. uvarum relative to S. cerevisiae). Note that Sh4 is plotted on a different scale. For specific coordinates of copy number variants, see table 1.