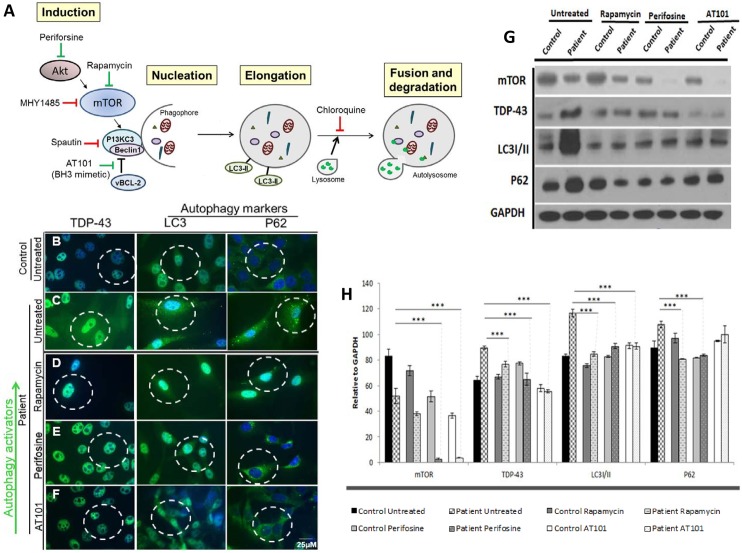
Fig 4. Drug screening with autophagy inducers Rapamycin, Perifosine and AT101 in patient VCP iPSC-derived myogenic lineages.
(A) Schematic of intervention with autophagy modifying agents. Green arrows show the active location of autophagy activators Rapamycin, Perifosine and AT101. Red arrows show the active location of autophagy inhibitors chloroquine, Spautin-1 and MHY1485. VCP is indicated to have an interactive role with Akt and as a chaperone protein with ubiquitin. (B) Untreated differentiated control and (C) untreated patient derived myogenic lineages. Patient derived myogenic lineage were treated with either (D) Rapamycin (10 μM), (E) Perifosine (80 μM) or (F) AT101 (10 μM) for 24 hours. Subsequently, cells were stained with TDP-43, LC3 or p62/SQSTM1 antibodies. Representative merged overlay images of stained iPSC with DAPI. Scale: Bar = 25 μm. White dotted lines represent areas of increased or decreased expressions. (G) Western blot analysis of iPSC-derived control (C) and patient (P) myoblasts with mTOR, TDP-43, LC3I/II and p62/SQSTM1. GAPDH was used as a positive loading control. (H) Densitometry analyses of the Western blot. Black dotted line indicates expression over baseline control sample. Statistical significance is denoted by *p<0.05, **p<0.005 and ***p<0.001.