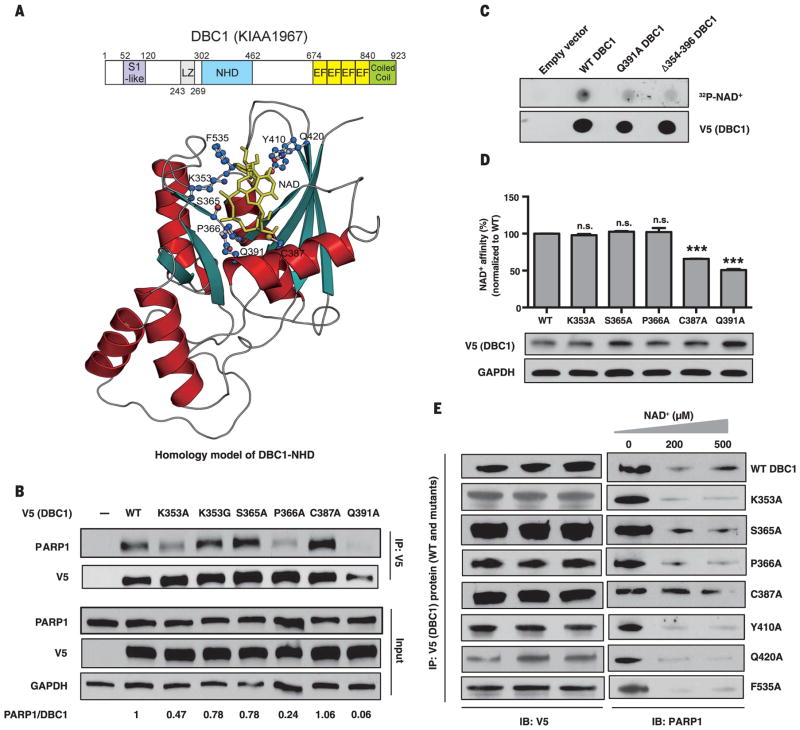
Fig. 2. Binding of the Nudix homology domain (NHD) of DBC1 to NAD+ and PARP1.
(A) Domains and crystallographic-based homology model of the NHD docked with NAD+. S1-like, ribosomal protein S1 OB-fold domain-like; EF, EF-hand; LZ, leucine zipper. Residues predicted to be in the vicinity of bound NAD+ are highlighted. (B) Interaction of V5/His-tagged DBC1 mutants and PARP1. See fig. S8 for additional mutants. (C and D) Direct binding of NAD+ to the DBC1-NHD, assessed using a radiolabeled NAD+ binding assay (C) or a biotin-NAD+ binding assay (D). Error bars indicate mean ± SEM; one-way ANOVA and Sidak’s post-hoc correction. ***P < 0.001; n.s., not significant. (E) Effect of NAD+ on binding of DBC1-NHD mutants to PARP1. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; F, Phe; G, Gly; K, Lys; P, Pro; Q, Gln; S, Ser; and Y, Tyr.