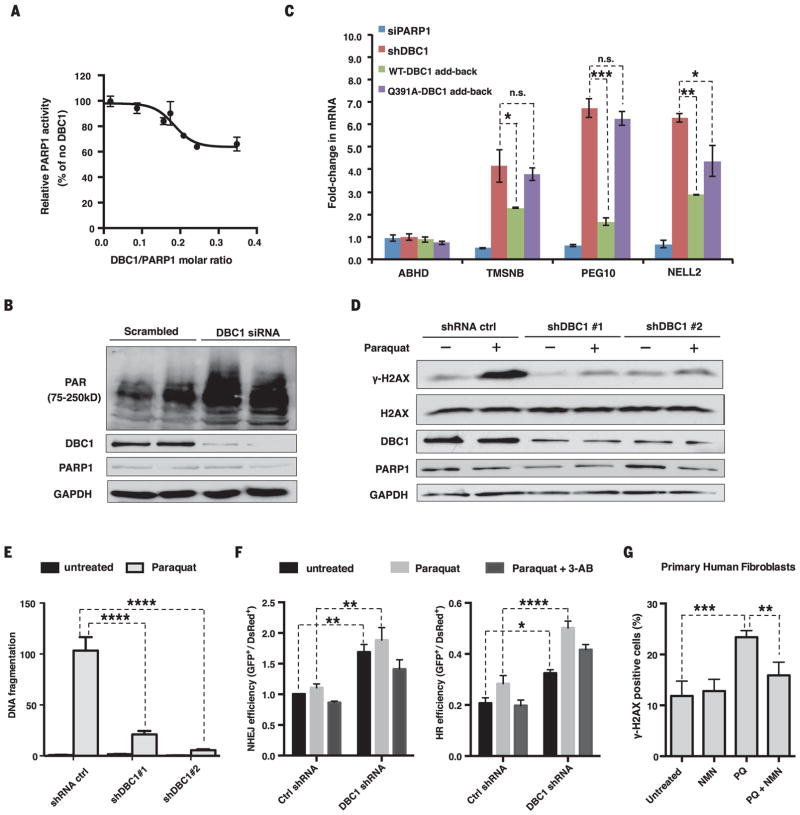
Fig. 3. DBC1 inhibits PARP1 activity and DNA repair.
(A) Inhibition of PARP1 activity by DBC1 purified from HEK 293T cells. (B) PAR [poly(ADP-ribose)] abundance in HEK 293T cells lacking DBC1. siRNA, small interfering RNA. (C) Opposing effects of reintroducing the wild type or DBC1Q391A into MCF-7 cells on mRNA of PARP1-regulated genes. TMSNB, Thymosin beta; PEG10, paternally expressed gene 10; NELL2, neural EGFL-like 2. ABHD2 was a negative control. DBC1 and PARP1 abundance are shown in fig. S10E. sh, short hairpin. (D) γH2AX abundance in DBC1 knockdown cells after paraquat treatment (1 mM, 24 hours). (E) DNA fragmentation after paraquat treatment (0.5 mM, 24 hours) in DBC1 knockdown cells, assessed by a comet assay (>50 cells per group). See fig. S11A. (F) DNA break repair [nonhomologous end joining (NHEJ) and homologous recombination (HR)] in DBC1 knockdown cells treated with paraquat (1 mM) or 3-AB (5 mM). n = 3 biological replicates. (G) Protection of human primary fibroblasts from paraquat (PQ)–induced DNA damage (300 μM paraquat) by NMN (500 μM). 100 ± 20 cells per condition, n = 4 biological replicates (two cell lines, twice for each), 24-hour treatment. See fig. S12. Error bars indicate SEM; one-way ANOVA [(C) and (G)] and two-way ANOVA [(E) and (F)], Sidak’s post-hoc correction. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n.s., not significant.