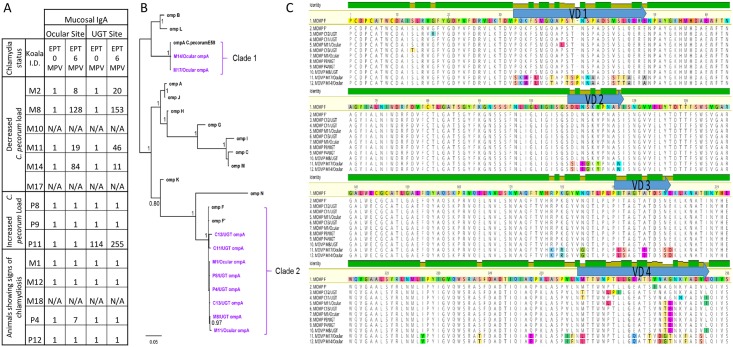
Fig 5. Comparison of chlamydia and disease status with mucosal IgA, alignment and phylogenetic analysis of the new ompA genotypes in infected koalas.
(A) Infection and disease status was assessed and compared with mucosal IgA in koalas pre- and post-vaccination. (B) Alignment of the new MOMP variants from strains infecting the koalas in the present vaccine trial including the previously described ompA F and F’ subtypes. (C) Bayesian phylogenetic analysis of the 24 ompA sequences, including the 10 koala C. pecorum ompA sequences generated in this study, and 13 previously described koala C. pecorum ompA genotypes. The C. pecorum E58 ompA sequence was used as an out-group. Posterior probabilities are displayed in the tree nodes.