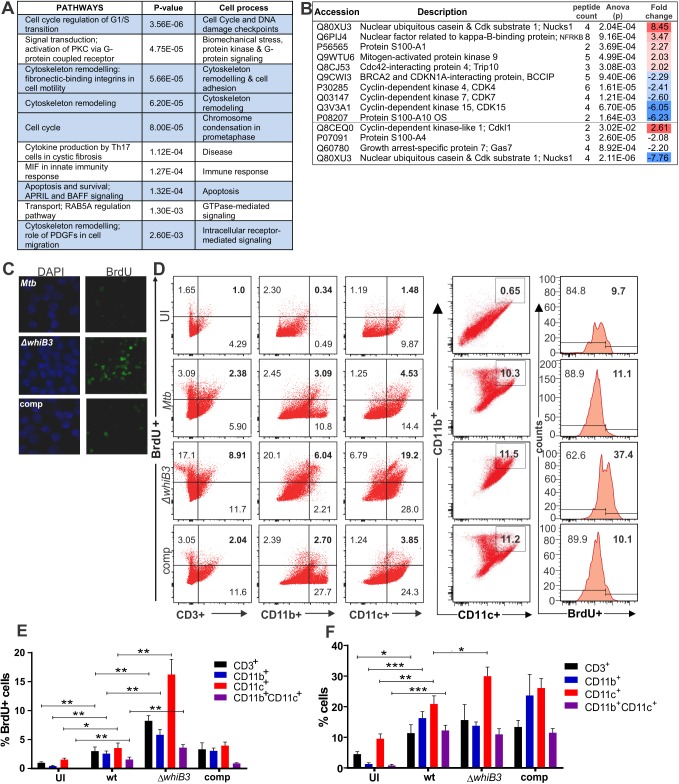
Fig 5. Pathway mapping of Mtb infected macrophages reveals that Mtb WhiB3 regulates the host cell cycle.
(A) Top ten host pathways with the most significant differential regulation in MtbΔwhiB3 infected RAW264.7 macrophages relative to wt Mtb H37Rv infected macrophages (p values were calculated with t tests). See S2 Fig and S5 and S6 Tables. (B) LC-MS/MS identification and quantification of cytoplasmic proteins with more than 2-fold differential expression (p<0.03, ANOVA) in MtbΔwhiB3 infected RAW264.7 macrophages relative to wt Mtb H37Rv infected macrophages. Fold changes were calculated from the means of samples in triplicate. See S7 Table. (C) DNA synthesis in RAW264.7 macrophages infected with wt Mtb H37Rv, MtbΔwhiB3 and the complemented (comp) strain (MOI 5) for 24 h. Cells were stained with DAPI (blue) to identify the nuclei and BrdU was added to the cells and incorporated into newly synthesized DNA. (D) Representative dot plots of BrdU incorporation versus surface staining with anti-CD3+ (lymphocyte marker), anti-CD11b+ and anti-CD11c+ (myeloid lineage markers) of lung cells isolated from mice infected with wt Mtb, MtbΔwhiB3 or the complemented strain for 6 weeks. CD11b+CD11c+ double positive cells were also gated and evaluated for BrdU incorporation. (E) Percentage of CD3+, CD11b+, CD11c+ or CD11b+CD11c+ cells isolated from mouse lungs infected with wt Mtb, MtbΔwhiB3 or the complemented strain that are positive for BrdU incorporation. (F) Percentage of total CD3+, CD11b+, CD11c+ or CD11b+CD11c+ cells isolated from mouse lungs infected with wt Mtb, MtbΔwhiB3 or the complemented strain. Data are representative of two independent experiments with three mice per group and were analyzed using the unpaired Student’s t test. For the comparison of MtbΔwhiB3 infected mice versus any of the other mouse groups: * p<0.01.