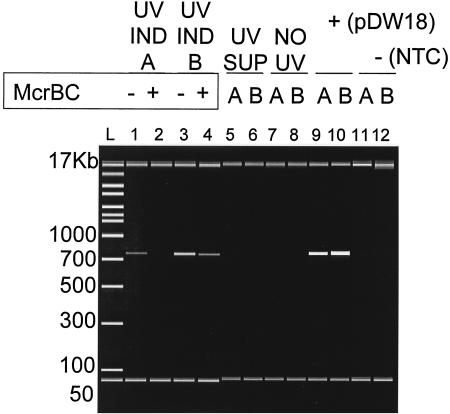
FIG. 3.
Induction of replication triggers methylated extrachromosomal phage accumulation in E125. Depicted is a representative digital gel output from an Agilent 2100 bioanalyzer using the DNA 12000 chip system. One microgram of genomic DNA from each of the indicated genomic DNAs was either digested with McrBC or mock digested under the NEB-suggested conditions. A total of ∼100 ng of each was then subjected to a limited PCR. Following PCR amplification using 15-s incubations, a 58°C annealing temperature, and 20 cycles, 1 μl of each 25-μl PCR mixture was loaded onto an Agilent 2100 bioanalyzer, which can size and quantify reaction products. Negative controls in this analysis were a water control (NTC) (lanes 11 and 12), any genomic DNA purified from the indicated cell supernatants (UV SUP) (lanes 5 and 6), and uninduced B. thailandensis E125 (NO UV) (lanes 7 and 8). The positive amplification control was a plasmid subclone (pDW18) that carries the HindIII fragment containing attP (lanes 9 and 10). Because these primers lie in the phage genome outside the tRNA gene carried by attP, the amplicon is phage specific and can detect the presence of only unintegrated phage genomes. Both BML10 and E125 were previously demonstrated to contain integrated ΦE125 at the genomic tRNA homologous to attP (22). Induced (actively replicating) phage genome methylation correlates with the presence of the unintegrated phage genomes. UV IND, UV induction.