Abstract
In the present report, we examined the responses of diabetic Goto-Kakizaki (GK) rats and control Wistar-Kyoto (WKY) rats fed either a standard chow or high-fat diet (HFD) from weaning to 20 weeks of age. This comparison included gene expression profiling of skeletal muscle using Affymetrix gene array chips. The expression profiling is interpreted within the context of a wide array of physiological measurements. Genes whose expressions are different between the 2 strains regardless of diet, as well as genes that differ between strains only with HFD, were identified. In addition, genes that were regulated by diet in 1 or both strains were identified. The results suggest that both strains respond to HFD by an increased capacity to oxidize lipid fuels in the musculature but that this adaptation occurs more rapidly in WKY rats. The results also demonstrated an impaired cytokine signalling and heightened inflammatory status in the GK rats.
Keywords: Diabetes, high-fat diet, skeletal muscle, gene expression, microarrays
Introduction
Type 2 diabetes mellitus (T2DM) is a heterogeneous collection of diseases with the single common characteristic of an impaired response to insulin. The loss of insulin response leads to hyperinsulinaemia and profound changes in both glucose and lipid metabolisms.1,2 The hypersecretion of insulin generally leads to β-cell failure and severe diabetes. Although there are novel examples in both humans and animals where the development of T2DM is associated with single gene defects, for the most part, the development of insulin resistance is caused by environmental factors acting on polygenic backgrounds. For example, studies as early as 1913 highlighted the relationship between the consumption of a high-fat diet (HFD) and the development of hyperglycaemia.3 The results presented here focus on the impact of high fat consumption on muscle gene expression in a polygenic model of T2DM.
The Goto-Kakizaki (GK) rat represents an animal model of T2DM with glucose intolerance, insulin resistance, and abnormal glucose metabolism.4,5 The insulin resistance causes hyperinsulinaemia which eventually leads to β-cell failure and the expression of chronic hyperglycaemia. This animal is a spontaneously diabetic rat produced by inbreeding of Wistar rats selected for high glucose values during oral glucose tolerance tests. However, diabetes in the GK rat is not associated with obesity. When fed a normal diet (ND), GK rats not only do not become obese but also actually exhibit an impaired development of mature adipocytes.6 Previous reports from our laboratory presented an extensive time-series analysis of skeletal muscle,7 liver,8 and adipose tissue6 from GK rats fed a normal rodent diet (10% fat) which demonstrated elevated chronic inflammation due to heightened natural immunity in all 3 tissues.
In additional studies, we compared those diabetic and control animals fed a normal rat diet from 4 to 20 weeks of age with a similar cohort fed a HFD (45% energy from fat) to examine the dietary effects on diabetes disease progression. Our analysis employed gene arrays along with extensive physiological measurements including body weights, organ weights, plasma glucose, plasma hormones (insulin, corticosterone, adiponectin, and leptin), and lipid profiles. Using the gene array data, we were able to identify genes whose expression is different between the 2 strains regardless of diet. In addition, we identified genes that responded to the HFD in both strains as well as those whose responses to HFD were unique to either GK or control animals. To date, we have published data on liver9 and adipose tissue10 from these studies. Because skeletal muscle is responsible for about 80% of insulin-directed glucose disposal, T2DM involves the impaired ability of the musculature to respond to insulin.11 This study presents gene array data from skeletal muscles taken from these animals.
Materials and Methods
Experimental design
A more extensive description of this experiment can be found in our published reports describing the array results on the livers and adipose tissue from these animals.9,10 Briefly, the experiment involved 25 GK rats spontaneously diabetic and 25 Wistar-Kyoto (WKY) nondiabetic male rats obtained from Taconic Farms (Germantown, NY, USA). They were maintained on a HFD (Harlan Teklad TD.06415 – 45% energy from fat) from 3 weeks of age until the end of the experiment. An additional set of 25 GK and 25 WKY animals fed standard rat chow (ND) were maintained and analysed in parallel. The research protocol adhered to the ‘Principles of Laboratory Animal Care’ (National Institutes of Health publication 85−23, revised in 1985) and was approved by the University at Buffalo Institutional Animal Care and Use Committee. Animals were maintained in our facility under environmental conditions with strict adherence to 12 hour:12 hour light:dark cycles. All manipulations and sacrifices were conducted between 1.5 and 3.5 hours after the beginning of the light cycle. Animals were housed in individual cages with free access to food and water. Food intake and body weights were measured twice weekly. Five animals from each strain were killed at 4, 8, 12, 16, and 20 weeks of age by aortic exsanguination using EDTA as anticoagulant. Animals were not fasted prior to sacrifice, and all measurements were performed following sacrifice. Gastrocnemius muscles were harvested, weighed, rapidly frozen in liquid nitrogen, and stored at −80°C. Comparison was made to identical groups of animals fed an ND (Harlan Teklad 2016 – 10% energy from fat).
Blood and plasma measurements
Glycosylated haemoglobin A1c (HbA1c) was measured using A1cNOW InView HbA1c test metres (Metrika, Sunnyvale, CA, USA) from whole blood taken at the time of sacrifice. Plasma glucose was measured by the glucose oxidase method (catalogue number GAGO-20; Sigma Chemicals, St. Louis, MO, USA) modified such that the assay was performed in a 1-mL assay volume with a 7-point standard curve. Plasma insulin was measured by a commercial enzyme-linked immunosorbent assay (ELISA) assay (Ultra Sensitive Rat Insulin ELISA kit; Crystal Chem Inc, Downers Grove, IL, USA). Plasma assays were conducted according to manufacturer’s directions with standards assayed in duplicate and experimental samples assayed in triplicate. Two experimental samples were selected as ‘quality controls’ for all assays to control possible interassay variations. Interand intra-assay variations were 10% or less. Individual values of plasma glucose and insulin were used to compute apparent homeostatic model assessment of insulin resistance (HOMA-IR) indices for each individual animal (HOMA-IR = glucose (mM) × insulin (µIU/mL)/22.5).12 A value of 44.45 µg/IU insulin was used for unit conversion. Values are reported as apparent HOMA-IR because animals were not fasted prior to sacrifice.
RNA preparation
Both gastrocnemius muscles from each animal were ground to a fine powder in a mortar cooled by liquid nitrogen, and tissue was added to prechilled TRIzol Lysis Reagent (Ambion, Carlsbad, CA, USA) in a weight/volume ratio of 1:10. Total RNA was extracted according to manufacturer’s directions and further purified using RNeasy mini columns (RNeasy Mini Kit; Qiagen Sciences, Germantown, MD, USA). Final RNA preparations were eluted in ribonuclease-free water, separated into aliquots and stored at −80°C. RNA was quantified spectrophotometrically and had 260/280 absorbance ratios of approximately 2.0. RNA samples were run on formaldehyde/agarose gel electrophoresis to assess purity and integrity. All samples showed intact ribosomal 28S and 18S RNA bands in an approximate ratio of 2:1.
Microarrays
Isolated RNA from each sample was used to prepare target according to manufacturer’s protocols. The biotinylated complementary RNAs (cRNAs) were hybridized to 50 individual Affymetrix GeneChips Rat Genome 230-2 (Affymetrix, Inc., Santa Clara, CA, USA) which contains 31 099 different probe sets.
Quantitative reverse transcription polymerase chain reaction
For validation purposes, gene-specific fluorescence-based real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays were developed and applied as previously described.13 The approach to quantitative RT-PCR involves use of in vitro–transcribed cRNA standards, gene-specific TaqMan-based probes, and a single-step assay with gene-specific messenger RNA (mRNA) normalized to total RNA in the assay.
Data mining
Affymetrix Microarray Suite 5.0 (Affymetrix) was employed for initial data acquisition and analysis. Signal intensities were normalized for each chip using a distribution of all genes around the 50th percentile. The generated data set is in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) database (GSE 13271). GeneSpring 7 (Silicon Genetics, Redwood City, CA, USA) was used for further analysis because it has a particularly useful approach for handling time-series data sets.
To objectively identify probe sets of interest, the entire data set was subjected to similar analyses procedures and filtered with the same criteria as those applied to previous gene array data sets for an identical experiment where the animals had received a normal rodent diet.6–8 Those previous results were presented as a comparison between GK-ND and WKY-ND animals. Here, with data sets from animals on HFD, 3 comparisons were made as follows: (1) GK-HFD vs GK-ND, (2) WKY-HFD vs WKY-ND, and (3) GK-HFD vs WKY-HFD. This approach does not select for probe sets but rather eliminates those probe sets that do not meet certain criteria, leaving the remainder for further consideration. In brief, the first filter eliminated genes not expressed in skeletal muscle, ie, those not having a call function of ‘P’ (present) in at least 5 of the 25 chips. The second level of filtering eliminated probe sets that could not meet the basic criterion of having a 2-fold expression difference between compared groups in at least 3 ages.
Statistics
For statistical comparisons, 2-way analyses of variance (ANOVAs) were conducted on raw or rank transformed data as appropriate using SigmaStat 3.5 software (Systat Software, Point Richmond, CA, USA) with Tukey post hoc tests.
Results
Postnatal growth and muscle development
Figure 1 shows the body weights and gastrocnemius muscle weights of the GK and WKY strains on both the ND and HFD diets as a function of age. Although the weight of both strains were similar at birth, by 8 weeks of age, the body weights of the WKY strain were significantly greater than the GK strain regardless of diet, a difference that remained throughout the entire 20-week period. What is interesting is that the effect of high-fat feeding was different on the body weights of the 2 strains. By 12 weeks of age, the GK strain on HFD was significantly heavier than the same strain on ND, whereas WKY on HFD was only marginally heavier than WKY on ND by 20 weeks. The increase in body weight was proportional to the increase in gastrocnemius muscle weight in GK animals, indicating that high-fat feeding promotes an increase in skeletal muscle in these animals. In contrast, in the WKY strain, an increase in muscle mass was only observed towards the end of the experiment.
Figure 1.
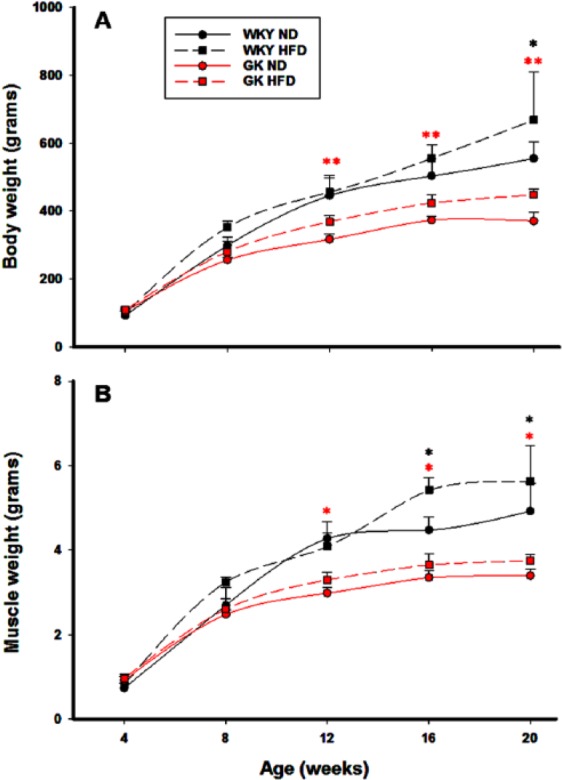
Growth characteristics of GK and WKY animals. (A) Total body weights as a function of age and (B) gastrocnemius muscle weights as a function of age. N = 5 animals per group. Symbols represent mean values and error bars 1 SD of the mean. Red asterisks reflect significant differences between GK-ND versus GK-HFD. Black asterisks reflect significant differences between WKY-ND versus WKY-HFD. *P < .05; **P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
Indices of insulin resistance
We previously published detailed reports on both the liver and adipose tissue from these animals on both diets. Data in those reports demonstrate that the GK strain had significantly higher plasma glucose and HbA1c than WKY (P < .001) from 4 weeks throughout the 20-week experimental period regardless of diet. Those data are presented in Supplemental Figure 1. However, because muscle is responsible for such a large part of insulin-directed glucose disposal, we used those data to calculate the apparent HOMA-IR indices for both strains receiving both diets. The apparent HOMA-IR (Figure 2) was significantly higher in GK animals compared with WKY animals from 8 weeks onwards, but diet did not have a significant effect on this index in either strain. This demonstrates that substantial insulin resistance was present in the GK strain from 8 weeks until the end of the experiment but was not apparently exacerbated by diet.
Figure 2.
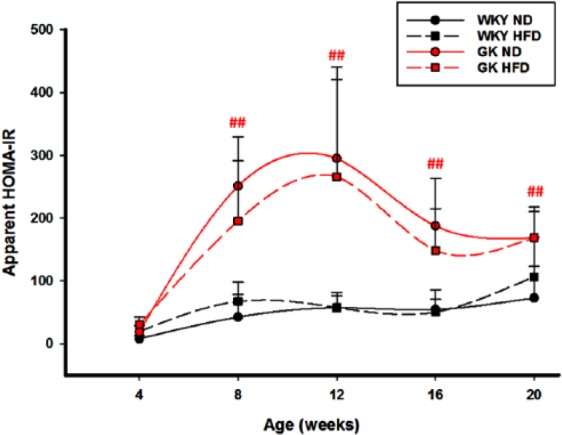
Indices of insulin resistance: Apparent HOMA-IR in GK and WKY rats with age. N = 5 animals per group. Symbols represent mean values and error bars 1 SD of the mean. No significant differences were apparent in HFD versus ND animals, but differences were significant between GK and WKY animals on either diet. ##P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
Data mining
Previously, we described gene array results from gastrocnemius muscles taken from both GK and WKY animals fed a standard rodent diet (ND – 10% fat).7 That report described a time series that was identical to the one used in this report and employed the identical data-mining approach. Having identical experimental time series from both strains on the 2 diets allowed us to address several sets of questions related to gene expression pertinent to understanding the difference between the 2 strains. These include genes that are differentially regulated by diet in only GK rats, genes differentially regulated by diet in only WKY rats, and genes differentially regulated by diet in both strains. In addition, these data allow us to compare genes that are differentially regulated in muscle from GK versus WKY animals fed HFD and to compare those differences between strains that occur when animals are fed an ND.
Genes responding to diet only in GK rats
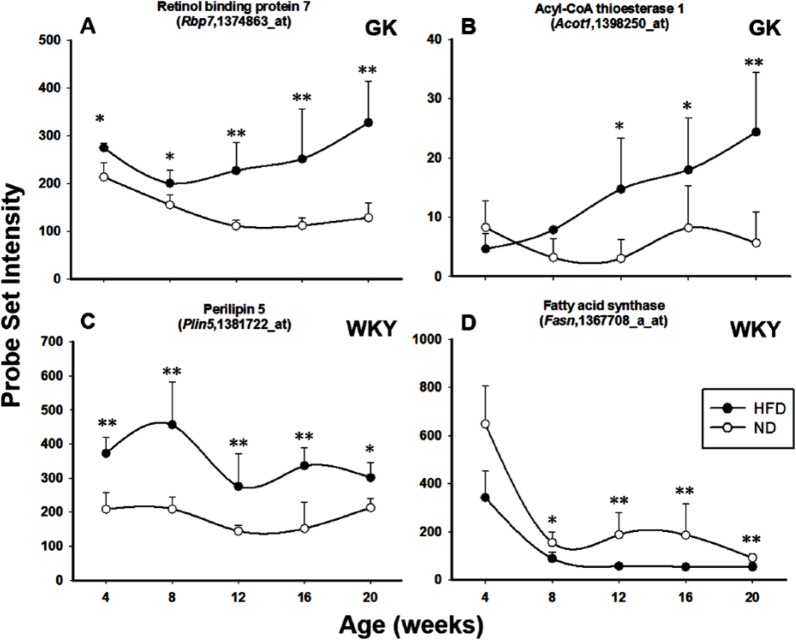
Supplemental Table 1 lists the genes that responded to HFD only in the GK strain and not in the WKY strain. This included only 9 identified genes. One relevant gene is retinol-binding protein 7, cellular (RBP7), also known as RBP4, CRBP4, or CRBP-III, which is a member of cellular retinol-binding protein family. It is the only specific transport protein for retinol (vitamin A) in the circulation and, to date, its only known function is to deliver retinol to tissues.14,15 Elevation of serum RBP7 has been causally linked to systemic insulin resistance through downregulation of GLUT4. High fat–fed RBP7-deficient mice have decreased serum free fatty acid (FFA) compared with high fat–fed wild-type mice15 and are protected against the development of insulin resistance.16 The deficiency of RBP7 was also associated with reduced food intake and altered body composition, with decreased adiposity and increased lean body mass. In addition, increased expression of genes involved in mitochondrial fatty acid oxidation in brown adipose tissue was observed in RBP7-deficient mice.15 RBP7 is more highly expressed in HFD-fed GK muscle than ND-fed GK muscle in all age groups (Figure 3A). In addition, its expression also somewhat increased with age with high-fat feeding, indicating the possibly elevated mitochondrial fatty acid oxidation in the HFD-fed GK muscle. Another pertinent gene is acyl-coenzyme A (CoA) thioesterase 1 (Acot1) which is involved in lipid metabolism by modulation of cellular concentrations of acyl-CoAs and fatty acids. Acot1 is a cytosolic acyl-CoA thioesterase which hydrolyses acyl-CoAs to the corresponding FFA and CoA. Acot1 is active on long-chain acyl-CoAs. One suggested role for Acot1 is in the control of ligand supply for the peroxisome proliferator–activated receptor (PPAR) family of nuclear receptors in the form of acyl-CoAs or FFAs and/or channelling fatty acids towards degradation rather than esterification.17,18 Acot1 has been shown to be increased in heart muscle from diabetic animals and has been associated with the development of insulin resistance.19,20 The expression level of Acot1 is elevated in GK-HFD muscle compared with GK-ND muscle from 8 to 20 weeks of age (Figure 3B). In addition, Acot1 expression increased with age in high fat–fed GK animals but not in ND-fed GKs.
Figure 3.
Genes responding to diet in only 1 strain. Examples of genes responding to diet in either GK rats only (A and B) or WKY rats only (C and D). Symbols represent mean values and error bars 1 SD of the mean. N = 5 animals per group. *P < .05; **P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
Genes responding to diet only in WKY rats
Supplemental Table 2 lists the 30 identified genes that responded to HFD only in the WKY strain. A number of genes indicate differentially regulated lipid metabolism in WKY skeletal muscle in response to HFD feeding compared with ND feeding. Perilipin 5 (Plin5) is a member of a family of proteins that coat the surfaces of intracellular neutral lipid storage droplets. In the basal state, Plin5 prevents access of hormone-sensitive lipase to the lipid droplets. Specific hormonal or cytokine stimuli, such as catecholamines and tumour necrosis factor α, activate lipolysis by phosphorylating perilipin; thereby allowing hormone-sensitive lipase to access the lipid droplet and initiate its lipolytic action.21 Plin5 is indicated to be important for intramuscular fat deposition.22,23 It has also been reported that Plin5-knockout mice developed skeletal muscle insulin resistance.24 Plin5 is approximately 2 times more highly expressed in WKY-HFD muscles compared with WKY-ND muscles, indicating adaptations in lipolysis to high-fat feeding (Figure 3C). Fatty acid synthase (Fasn) is also differentially regulated by diet in WKY muscle. Fasn encodes a lipogenic enzyme whose main function is to catalyse the synthesis of palmitate from acetyl-CoA and malonyl-CoA in the presence of reduced nicotinamide adenine dinucleotide phosphate into long-chain saturated fatty acids.25 Deletion of Fasn in macrophages has been shown to prevent diet-induced insulin resistance.26 Fasn has a higher expression in WKY-ND muscle than WKY-HFD muscle from 8 weeks onwards, suggesting decreased lipogenesis in muscle with high-fat feeding (Figure 3D).
Genes responding to diet in both strains
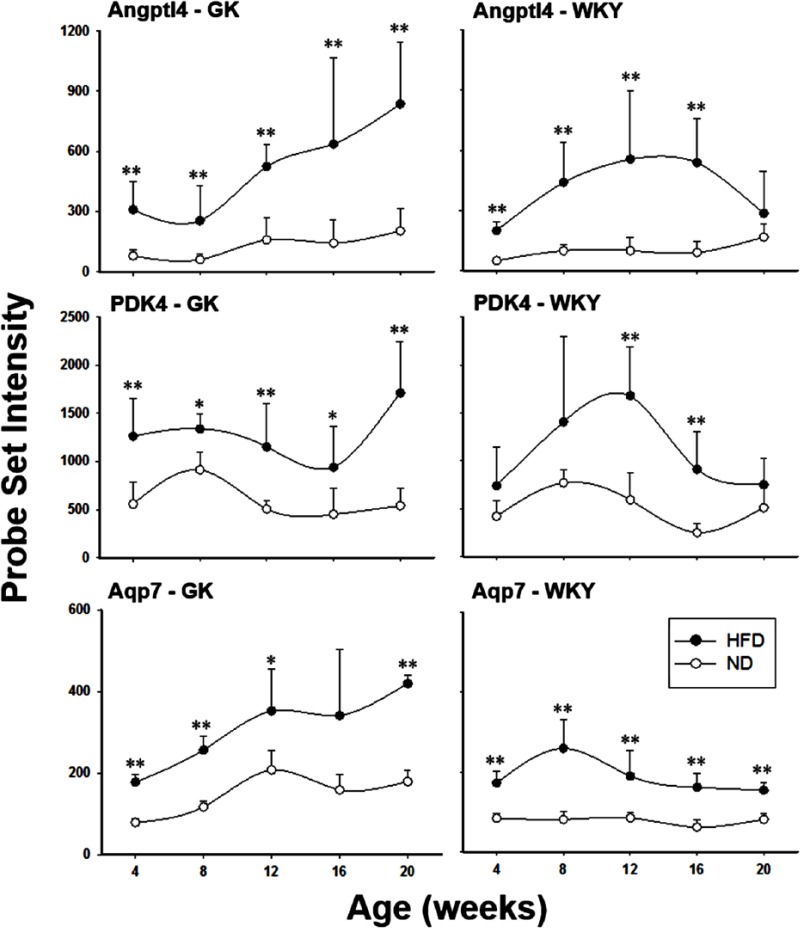
Supplemental Table 3 lists 8 additional identified genes that responded to HFD in both strains. The predominant source of energy for resting skeletal muscle is fatty acid oxidation. Glucose and ketone body metabolisms also contribute to different degrees, depending on the activity and metabolic state of the muscle. Switching to a HFD has been shown to lead to increased fluxes of fatty acids into muscle tissue.27 Thus, it is not surprising that there is a significant difference between HFD-fed and ND-fed rats in the mRNAs for proteins involved in energy metabolism. For example, angiopoietin-like 4 (Angptl4), a secreted protein that has been demonstrated to regulate triglyceride metabolism by inhibiting lipoprotein lipase (LPL),28 has higher expression both in high fat–fed GK and WKY rats compared with ND-fed animals of the same strain (Figure 4 – top panels), although there is no significant difference between high fat–fed GK and high fat–fed WKY animals. Lipoprotein lipase is an endothelium-associated enzyme that hydrolyses the triacylglycerol component of circulating chylomicrons and very low-density lipoproteins, resulting in the production of non-esterified fatty acids and 2-monoacylglycerol for tissue use. Previous research also indicates that administration of recombinant Angptl4 protein to mice mediates hypertriglyceridaemia, possibly through inhibition of LPL.29,30 In addition, pyruvate dehydrogenase kinase isozyme 4 (PDK4) is differentially regulated by diet in both strains (Figure 4 – middle panels). PDK4 mediates pyruvate oxidation through inhibitory phosphorylation of the pyruvate dehydrogenase complex. There is a positive correlation between the increase in PDK4 expression and the tendency to use lipid-derived fuels as respiratory substrates in muscle.31 Increased expression of PDK4 has been linked to skeletal muscle insulin resistance,32 and PDK4 has been identified as a candidate gene for T2DM.33 In our study, the HFD enhanced PDK4 expression in both GK and WKY rats and reflects the elevated contribution of fatty acid as an energy source for muscle in response to high-fat feeding. This conclusion is further supported by the observation of higher expression of aquaporin 7 (Aqp7) in the muscle from high fat–fed rats (Figure 4 – bottom panels). Aqp7 is an aquaglyceroporin responsible for glycerol efflux from skeletal muscle.34 The higher expression of Aqp7 at all age groups in both strains suggests a higher level of lipolysis in these muscles. Increased expression of this gene has also been demonstrated in skeletal muscle from ob/ob mice.35
Figure 4.
Genes responding to diet in both strains. Examples of differentially regulated genes responding to diet in both GK (left panels) and WKY (right panels) animals. Symbols represent mean values and error bars 1 SD of the mean. N = 5 animals per group. *P < .05; **P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
Effects of diet on gene expression differences between strains
Based on our filtering approach, there were 307 probe sets that were differentially expressed in skeletal muscle comparing GK-HFD and WKY-HFD animals. Previously, using the same mining approach, we identified 318 probe sets that were differentially expressed in animals fed a normal rodent diet.6 As illustrated in Figure 5, 215 of the probe sets were differentially expressed between strains on either diet (green), whereas 92 were unique to HFD-fed animals (yellow). These 307 probe sets are listed in Supplemental Table 4, with the 92 probe sets differentially regulated only with HFD highlighted in bold font. Of the 318 differentially regulated probe sets from ND-fed animals, 103 of these were not differentially expressed in HFD rats (Figure 5, blue). These probe sets are listed in Supplemental Table 5.
Figure 5.
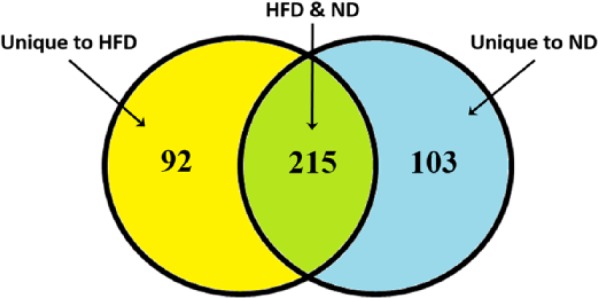
Venn diagram of number of probe sets differentially regulated between GK and WKY animals as a function of diet. Yellow, different between GK and WKY only on HFD; green, different between GK and WKY on either diet; and blue, different between GK and WKY only on ND. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
HFD-dependent differences in gene expression between GK and WKY rats
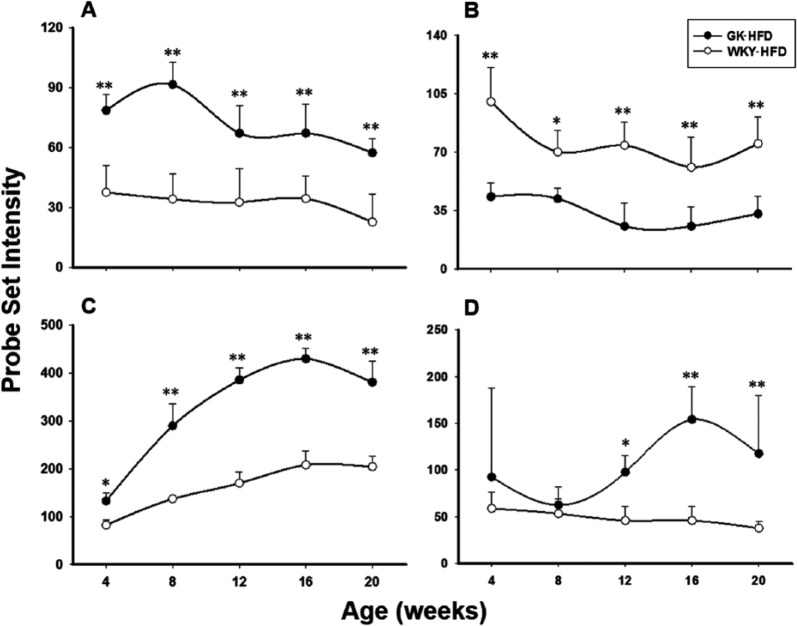
Among these were several genes involved in immune function. For example, proteasome (prosome, macropain) subunit, β type-8 (Psmb8) is a member of the proteasome B-type family whose function is to generate class I major histocompatibility complex (MHC) peptides. It is also implicated in the autoimmune basis in type 1 diabetes.36 Psmb8 is approximately 2 times higher in GK-HFD rats compared with WKY-HFD at all age groups, indicating an elevated immune response in the diabetic GK rats fed a HFD (Figure 6A). This conclusion is further supported by the reduced inositol hexakisphosphate kinase 2 (IP6K2) expression in muscles from high fat–fed GK animals compared with high fat–fed WKY animals (Figure 6B). IP6K2 is a kinase that catalyses the synthesis of diphosphoinositol pentakisphosphate and bis-diphosphoinositol tetrakisphosphate. IP6K2 is known as a proapoptotic gene. Previous studies indicated an association of IP6K2 with cell death.37 Overexpression of IP6K2 increases the sensitivity of cancer cell lines to apoptotic actions of cell stressors, whereas depletion of IP6K2 by RNA interference prevents the apoptotic actions. Recently, IHPK2 has been demonstrated to bind to tumour necrosis factor receptor–associated factor 2 and interferes with phosphorylation of transforming growth factor β–activated kinase 1 (TAK1), thereby inhibiting nuclear factor κB (NF-κB) signalling.38 The expression level of IP6K2 was reduced in GK-HFD muscles compared with WKY-HFD muscles at all age groups. Expression of Slc4a4 is presented in Figure 6C. Solute carrier family 4 (anion exchanger), member 4 (slc4a4) encodes a sodium bicarbonate cotransporter (NBC) involved in the regulation of bicarbonate secretion and absorption and intracellular pH.39 Muscle bicarbonate content was related to the degree of metabolic acidosis which often leads to loss of body protein due mainly to accelerated protein breakdown in muscle.40 At 4 weeks of age, Slc4a4 mRNA was only slightly higher in GK-HFD compared with WKY-HFD muscles. With increasing age, this difference increases such that by 20 weeks of age it is about 2-fold higher in GK-HFD compared with WKY-HFD. This might suggest a potential increase in muscle protein catabolism, which is also observed in type 1 diabetes.41 Tia1, cytotoxic granule–associated RNA binding protein–like 1 (Figure 6D) is a member of a family of RNA-binding proteins which functions as a posttranscriptional regulator of gene expression and forms stress granules following cellular damage.42 Tial1 mRNA expression is relatively constant in muscle from WKY-HFD animals. However, in GK-HFD muscle, its expression level increases with age, and from 12 weeks of age onwards, there is higher expression in GK animals compared with WKY, indicating possible muscle damage in the GK diabetic rats.
Figure 6.
HFD-dependent differences in gene expression between GK and WKY rats. Examples of differentially expressed genes between GK-HFD and WKY-HFD but not different when animals are fed a normal diet. (A) Psmb8, (B) Ip6k2, (C) Slc4a4, and (D) Tial1. Symbols represent mean values and error bars 1 SD of the mean. N = 5 animals per group. *P < .05; **P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; WKY, Wistar-Kyoto.
Diet-independent differences in gene expression between GK and WKY rats
A number of genes were differentially expressed between diabetic GK and normoglycaemic WKY animals regardless of diet. Figure 7A shows the expression of iron-sulfur cluster assembly 1 homologue (Isca1) in muscle from both populations. Isca1 is a nuclear-encoded mitochondrial gene involved in the assembling of mitochondrial iron-sulfur proteins and plays an important role in electron transport.43 The expression of Isca1 is severalfold higher at all ages in GK relative to WKY regardless of diet which suggests that there are more mitochondria in the muscles of the diabetic animals. Similarly, the higher expression of mitochondrial ribosomal protein L52 (Mrpl52) in GK samples in both the ND and HFD also suggests that there are more mitochondria in the muscles of the GK population (Figure 7B). Mrpl52, encoded by a nuclear gene, is involved in protein synthesis within the mitochondrion. It is a component of the mitochondrial ribosome large subunit (39S) which comprises a 16S ribosomal RNA and about 50 distinct proteins.44 There are also a number of genes that indicate a heightened inflammatory state in the musculature of the GK animals regardless of diet. Interferon-induced protein with tetratricopeptide repeats 1 (Ifit1) which is associated with chronic inflammation and insulin resistance is substantially higher at all ages in the GK muscle regardless of diet (Figure 7C). Interferons are a family of proteins that respond to the presence of pathogens such as viruses, bacteria, parasites, and tumour cells. Interferon is involved in natural anti-virus immunity. When activated by the immune system, interferon signals neighbouring cells and triggers their resistance mechanisms. In addition, it activates other immune cells which kill invading pathogens.45,46 A comparison of Ifit1 expression measured by both gene arrays and by quantitative qRT-PCR is presented in Supplemental Figure 2, demonstrating that the pattern of expression is virtually identical with the 2 methodologies. The expression of suppressor of cytokine signalling 2 (Socs2), which is a negative regulator of cytokine signalling, is higher in the muscles from the WKY population regardless of diet (Figure 7D). The SOCS family proteins form part of a classical negative feedback system that regulates cytokine signal transduction.47 Socs2 appears to be a negative regulator in the growth hormone/IGF1 signalling pathway.48 It is induced by a subset of cytokines, including erythropoietin and granulocyte-macrophage colony-stimulating factor. It is also involved in the interleukin 6–induced receptor phosphorylation and in the negative regulation of cytokine-induced STAT factor signalling.49 When fed a normal rodent diet, the Socs2 expression level remained relative constant in the GK rats, whereas in the control WKY rats, it increased sharply between 4 and 8 weeks and slowly declined through the rest of experiment period but was still significantly higher at all age groups compared with the diabetic GK rats. In the HFD animals, there was a moderate increase between 4 and 8 weeks in GK rats, then quickly returned to initial level at 12 weeks and remained at that level. However, in WKY rats, there was an abrupt increase between 4 and 8 weeks and remained elevated with only a slight decline at 20 weeks old. This result suggests an impaired cytokine signalling pathway and possible heightened inflammation status in the diabetic GK rats.
Figure 7.
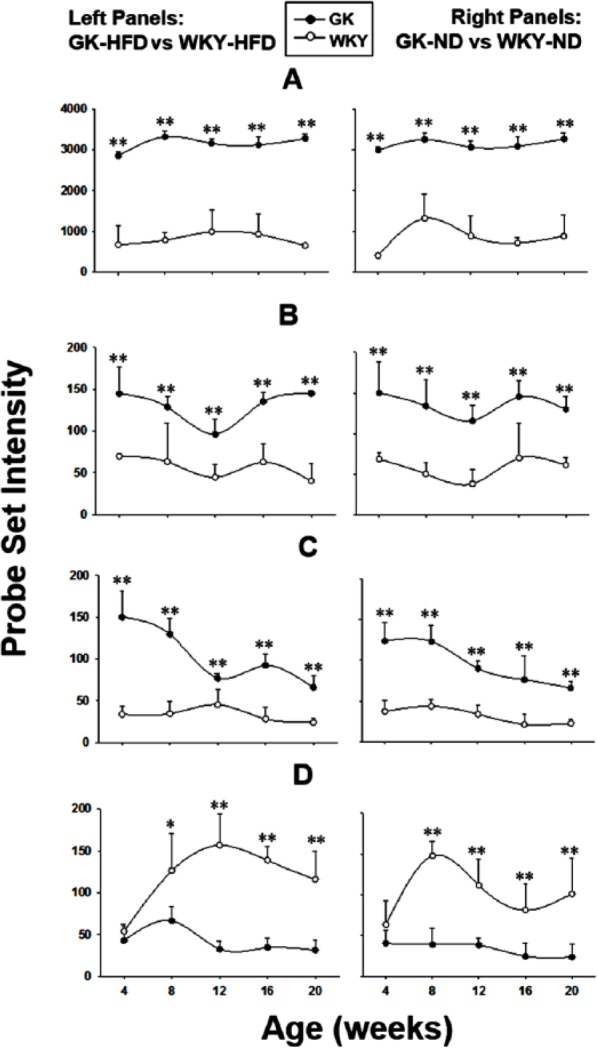
Diet-independent differences in gene expression between GK and WKY rats. Examples of genes whose expression differs between GK and WKY animals, both when fed HFD (left panels) and ND (right panels). (A) Isca1, (B) Mrpl52, (C) Ifit1, and (D) Socs2. Symbols represent mean values and error bars 1 SD of the mean. N = 5 animals per group. *P < .05; **P < .001. GK indicates Goto-Kakizaki; HFD, high-fat diet; ND, normal diet; WKY, Wistar-Kyoto.
Discussion
Insulin-resistant diabetes, T2DM, to a great degree, must involve the musculature because collectively this tissue is responsible for about 70% to 80% of the insulin-directed glucose disposal. A HFD is a major risk factor in the development of T2DM. One element of the relationship of high-fat intake to the development of T2DM is the accumulation of excess adipose tissue which in turn causes chronic inflammation due to the invasion of macrophages.50 Chronic inflammation impairs the ability of the musculature and other tissues to respond to insulin. A second element in the relationship of fat intake to the development of T2DM is that such a diet provides an abundance of lipid fuels that reduces the reliance of the musculature on glucose.
Previously, we conducted extensive comparative analyses of diabetic GK and normoglycaemic WKY rats fed a normal rodent diet (10% energy from fat). Those studies provided strong evidence for the hypothesis that there was chronic inflammation in the GK animals mediated by an elevated natural immunity. Such heightened natural immunity was indicated by an extensive gene expression analysis of liver, skeletal muscle, and adipose from the GK strain. That time-series experiment also demonstrated that the diabetic GK strain was deficient in mature adipocytes.6 This hypothesis was further tested by chronic treatment of the GK strain with salsalate which blocks the activation of NF-κB. The results showed that salsalate greatly reduced the hyperglycaemia normally observed in the GK strain.13
As a follow-up to the ND time series, we conducted an identical experiment except that both strains were fed a HFD (45% energy from fat). The HFD had only marginal effect on indices of diabetes in the GK strain (Supplemental Figure 1). Specifically, development of diabetes as indicated by increased plasma glucose was delayed by about 4 weeks with high-fat feeding. In contrast, the WKY strain became hyperinsulinaemic after 16 weeks suggesting that the controls became prediabetic towards the end of the experiment. Interestingly, HFD significantly mitigated the adipose tissue deficiency in the GK strain noted when the animals were fed an ND.
In the present report, we evaluated the effect of HFD on the musculature of both strains. High-fat diet had only marginal effects on total body weight in the WKY strain and an increase in relative muscle mass was only noted at 20 weeks. In contrast, HFD caused a significant increase in body weight in the GK strain (Figure 1). What is interesting is that there is a proportional increase in muscle mass in the GK strain. This result suggests that high-fat feeding promotes the development of the musculature in the GK animals. The most plausible explanation is that on the ND, the insulin-resistant musculature of the GK strain is starved for energy impeding muscle development, whereas the HFD provided an alternative fuel for the musculature thereby promoting its development. This hypothesis is supported by the observation that there is a significant increase in muscle triglyceride content by 12 weeks in the GK strain (data not shown), whereas no such increase is noted in the WKY strain.
To provide context for observed changes, we used gene arrays to evaluate gene expression effects of HFD on both strains. We used the gene expression data to conduct a within strain and cross-strain analysis with the same time-series progression when the animals were fed an ND. A group of genes indicate muscle adaptation to HFD through elevated fat oxidation in the muscle. For example, the expression of PDK4 is higher at all age groups in both strains with high-fat feeding (Figure 4). This may reflect an increased capacity to oxidize lipid fuels in the high fat–fed musculature, possibly due to the excess influx of fatty acid from the diet. This conclusion is further supported by the observation that expression of Plin5, which mediates access of hormone-sensitive lipase to the lipid droplets, is higher in the WKY-HFD muscles (Figure 3). In addition, the expression of Fasn, a lipogenic enzyme, is lower at all age groups in the WKY-HFD muscles, indicating a lower level of lipogenesis (Figure 3). However, the expression of these genes is not significantly different between GK and WKY animals fed the same diet. Therefore, these changes in WKY muscle imply that muscle adapts to the diet with increased ability to oxidize fat, but does not indicate insulin resistance development. The expressions of Aqp7, PDK4, and Angptl4, which are differentially regulated in response to HFD in both strains, are higher at all age groups in the high fat–fed GK animals (Figure 4). However, in WKY animals, the elevated expression in HFD animals tends to decline at later ages, suggesting the development of tolerance.
Circulating FFAs are derived from lipolysis in adipose tissue, and some evidence suggests that these FFAs provide excess lipid fuel for the musculature, reducing glucose uptake. The high fat–fed animals have significantly higher plasma FFA levels than the ND-fed animals.9 However, consistent with previous studies, only GK rats, which have a genetic predisposition to insulin resistance and diabetes, have higher muscle triglyceride contents after high-fat feeding, indicating metabolic inflexibility within the skeletal muscle in response to dietary lipid challenge. This conclusion is further supported by the microarray data. One major mechanism which skeletal muscle uses to adapt to a HFD is through ligand activation of the PPARα transcription factor.51,52 Peroxisome proliferator–activated receptor α activation leads to transcriptional activation of multiple enzymes in the pathways of fatty acid utilization. However, there is also evidence indicating that inappropriate activation of PPARα in the muscle can be deleterious during a high saturated fat diet.53 Acot1 is involved in ligand supply for the PPAR family of nuclear receptors and has higher expression in HFD GK muscle compared with ND GK muscle from 8 to 20 weeks of age (Figure 3). In addition, GK-HFD muscle has significantly higher Acot1 expression than WKY-HFD, but no difference was observed between GK-ND and WKY-ND. Moreover, unlike in WKY muscles, which quickly adapts to HFD, the expressions of Aqp7, PDK4, and Angptl4 are continuously elevated as age increases (Figure 4). RBP7, which is linked to causing systemic insulin resistance through downregulation of GLUT4 and increasing the mitochondrial fatty acid oxidation, has higher expression at all age groups in HFD-fed GKs, but no diet effects were observed in WKYs (Figure 3). Regardless of diet, there are indications of more mitochondria in diabetic GK rats. The expression levels of the mitochondria proteins Isca1 as well as Mrpl52 are elevated in GK muscle at all age groups (Figure 7). This may reflect an increased capacity to oxidize lipid fuels in the GK musculature, which may be a compensation for a reduced capacity to bring in glucose.
Increased inflammation is also evident in GK animals regardless of diet. The higher expression of Ifit1 and the lower expression of Socs2 in GK muscle compared with WKY controls regardless of diet indicate impaired cytokine signalling and heightened inflammatory status in the GK rats (Figure 7). This conclusion is further supported by observations that the expression level of Ip6k2 which interferes with phosphorylation of TAK1 and inhibits NF-κB signalling is higher in the WKY-HFD muscles compared with those from high fat–fed GK rats, whereas the expression of Psmb8 which generates class I MHC peptides is elevated in GK-HFD compared with WKY-HFD muscles (Figure 6).
Conclusions
Our results indicated heightened inflammatory status and increased mitochondria content/activity in the skeletal muscle of GK rats. The combination of these metabolic derangements, together with slower adaptation to the HFD, manifests an underlying disorder intrinsic to skeletal muscle that contributes to the dietary effects on diabetic GK rats.
Supplementary Materials
Acknowledgments
The authors would like to thank Ms Nancy Pyszczynski for expert technical assistance.
Footnotes
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partly supported by grant GM 24211 from the National Institute of General Medical Sciences, NIH, Bethesda, MD, and by funds from the UB-Pfizer Strategic Alliance.
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totalled 2764 words, excluding any confidential comments to the academic editor.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
DCD, WJJ, and RRA conceived and designed the experiments. JN, BX, DCD, and RRA analysed the data. JN wrote the first draft of the manuscript. JN, DCD, BX, WJJ, and RRA contributed to the writing of the manuscript; agree with manuscript results and conclusions; jointly developed the structure and arguments for the paper; and made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
Disclosures and Ethics
As a requirement of publication, author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material.
REFERENCES
- 1.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Bruce-Keller AJ, Dasuri K, Nguyen AT, Liu Y, Keller JN. Diet-induced metabolic disturbances as modulators of brain homeostasis. Biochim Biophys Acta. 2009;1792:417–422. doi: 10.1016/j.bbadis.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akash MS, Rehman K, Chen S. Goto-Kakizaki rats: its suitability as non-obese diabetic animal model for spontaneous type 2 diabetes mellitus. Curr Diabetes Rev. 2013;9:387–396. doi: 10.2174/15733998113099990069. [DOI] [PubMed] [Google Scholar]
- 5.Portha B, Giroix MH, Tourrel-Cuzin C, Le-Stunff H, Movassat J. The GK rat: a prototype for the study of non-overweight type 2 diabetes. Methods Mol Biol. 2012;933:125–159. doi: 10.1007/978-1-62703-068-7_9. [DOI] [PubMed] [Google Scholar]
- 6.Xue B, Sukumaran S, Nie J, Jusko WJ, Dubois DC, Almon RR. Adipose tissue deficiency and chronic inflammation in diabetic Goto-Kakizaki rats. PLoS ONE. 2011;6:e17386. doi: 10.1371/journal.pone.0017386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie J, Xue B, Sukumaran S, Jusko WJ, Dubois DC, Almon RR. Differential muscle gene expression as a function of disease progression in Goto-Kakizaki diabetic rats. Mol Cell Endocrinol. 2011;338:10–17. doi: 10.1016/j.mce.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almon RR, DuBois DC, Lai W, Xue B, Nie J, Jusko WJ. Gene expression analysis of hepatic roles in cause and development of diabetes in Goto-Kakizaki rats. J Endocrinol. 2009;200:331–346. doi: 10.1677/JOE-08-0404. [DOI] [PubMed] [Google Scholar]
- 9.Almon RR, Dubois DC, Sukumaran S, et al. Effects of high fat feeding on liver gene expression in diabetic Goto-Kakizaki rats. Gene Regul Syst Bio. 2012;6:151–168. doi: 10.4137/GRSB.S10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue B, Nie J, Wang X, DuBois DC, Jusko WJ, Almon RR. Effects of high fat feeding on adipose tissue gene expression in diabetic Goto-Kakizaki rats. Gene Regul Syst Bio. 2015;9:15–26. doi: 10.4137/GRSB.S25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koistinen HA, Zierath JR. Regulation of glucose transport in human skeletal muscle. Ann Med. 2002;34:410–418. doi: 10.1080/078538902321012351. [DOI] [PubMed] [Google Scholar]
- 12.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, DuBois DC, Cao Y, Jusko WJ, Almon RR. Diabetes disease progression in Goto-Kakizaki rats: effects of salsalate treatment. Diabetes Metab Syndr Obes. 2014;7:381–389. doi: 10.2147/DMSO.S65818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham TE, Yang Q, Bluher M, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–2563. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 15.Zizola CF, Schwartz GJ, Vogel S. Cellular retinol-binding protein type III is a PPARgamma target gene and plays a role in lipid metabolism. Am J Physiol Endocrinol Metab. 2008;295:E1358–E1368. doi: 10.1152/ajpendo.90464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry DC, Jacobs H, Marwarha G, et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 2013;288:24528–24539. doi: 10.1074/jbc.M113.484014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt MC, Nousiainen SE, Huttunen MK, Orii KE, Svensson LT, Alexson SE. Peroxisome proliferator-induced long chain acyl-CoA thioesterases comprise a highly conserved novel multi-gene family involved in lipid metabolism. J Biol Chem. 1999;274:34317–34326. doi: 10.1074/jbc.274.48.34317. [DOI] [PubMed] [Google Scholar]
- 18.Hunt MC, Rautanen A, Westin MA, Svensson LT, Alexson SE. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Calvo R, Vazquez-Carrera M, Masana L, Neumann D. AICAR protects against high palmitate/high insulin-induced intramyocellular lipid accumulation and insulin resistance in HL-1 cardiac cells by inducing PPAR-target gene expression. PPAR Res. 2015;2015:785783. doi: 10.1155/2015/785783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S, Chen C, Wang H, et al. Protective effects of acyl-coA thioesterase 1 on diabetic heart via PPARα/PGC1α signaling. PLoS ONE. 2012;7:e50376. doi: 10.1371/journal.pone.0050376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Souza SC, Muliro KV, Liscum L, et al. Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J Biol Chem. 2002;277:8267–8272. doi: 10.1074/jbc.M108329200. [DOI] [PubMed] [Google Scholar]
- 22.Laurens C, Bourlier V, Mairal A, et al. Perilipin 5 fine-tunes lipid oxidation to metabolic demand and protects against lipotoxicity in skeletal muscle. Sci Rep. 2016;6:38310. doi: 10.1038/srep38310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu JX, Albrecht E, Viergutz T, Nurnberg G, Zhao RQ, Wegner J. Perilipin, C/EBPalpha, and C/EBPbeta mRNA abundance in longissimus muscle and different adipose tissues of Holstein and Charolais cattle. Meat Sci. 2009;83:120–126. doi: 10.1016/j.meatsci.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Mason RR, Mokhtar R, Matzaris M, et al. PLIN5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Mol Metab. 2014;3:652–663. doi: 10.1016/j.molmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusunoki J, Kanatani A, Moller DE. Modulation of fatty acid metabolism as a potential approach to the treatment of obesity and the metabolic syndrome. Endocrine. 2006;29:91–100. doi: 10.1385/ENDO:29:1:91. [DOI] [PubMed] [Google Scholar]
- 26.Wei X, Song H, Yin L, et al. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature. 2016;539:294–298. doi: 10.1038/nature20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riccardi G, Giacco R, Rivellese AA. Dietary fat, insulin sensitivity and the metabolic syndrome. Clin Nutr. 2004;23:447–456. doi: 10.1016/j.clnu.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Koster A, Chao YB, Mosior M, et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 29.Koishi R, Ando Y, Ono M, et al. Angptl3 regulates lipid metabolism in mice. Nat Genet. 2002;30:151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida K, Shimizugawa T, Ono M, Furukawa H. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res. 2002;43:1770–1772. doi: 10.1194/jlr.c200010-jlr200. [DOI] [PubMed] [Google Scholar]
- 31.Holness MJ, Kraus A, Harris RA, Sugden MC. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes. 2000;49:775–781. doi: 10.2337/diabetes.49.5.775. [DOI] [PubMed] [Google Scholar]
- 32.Rinnankoski-Tuikka R, Silvennoinen M, Torvinen S, et al. Effects of high-fat diet and physical activity on pyruvate dehydrogenase kinase-4 in mouse skeletal muscle. Nutr Metab (Lond) 2012;9:53. doi: 10.1186/1743-7075-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasche A, Al-Hasani H, Herwig R. Meta-analysis approach identifies candidate genes and associated molecular networks for type-2 diabetes mellitus. BMC Genomics. 2008;9:310. doi: 10.1186/1471-2164-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Yu QQ, Zhu YH, et al. Insulin therapy stimulates lipid synthesis and improves endocrine functions of adipocytes in dietary obese C57BL/6 mice. Acta Pharmacol Sin. 2010;31:341–346. doi: 10.1038/aps.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakayama Y, Hirako S, Ogawa T, Jimi T, Shioda S. Upregulated expression of AQP 7 in the skeletal muscles of obese ob/ob mice. Acta Histochem Cytochem. 2014;47:27–33. doi: 10.1267/ahc.13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atkinson MA. ADA Outstanding Scientific Achievement Lecture 2004. Thirty years of investigating the autoimmune basis for type 1 diabetes: why can’t we prevent or reverse this disease? Diabetes. 2005;54:1253–1263. doi: 10.2337/diabetes.54.5.1253. [DOI] [PubMed] [Google Scholar]
- 37.Chakraborty A, Koldobskiy MA, Sixt KM, et al. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison BH, Bauer JA, Lupica JA, et al. Effect of inositol hexakisphosphate kinase 2 on transforming growth factor beta-activated kinase 1 and NF-kappaB activation. J Biol Chem. 2007;282:15349–15356. doi: 10.1074/jbc.M700156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero MF, Boron WF. Electrogenic Na+/HCO3− cotransporters: cloning and physiology. Annu Rev Physiol. 1999;61:699–723. doi: 10.1146/annurev.physiol.61.1.699. [DOI] [PubMed] [Google Scholar]
- 40.Arieff AI. Indications for use of bicarbonate in patients with metabolic acidosis. Br J Anaesth. 1991;67:165–177. doi: 10.1093/bja/67.2.165. [DOI] [PubMed] [Google Scholar]
- 41.Charlton M, Nair KS. Protein metabolism in insulin-dependent diabetes mellitus. J Nutr. 1998;128:323S–327S. doi: 10.1093/jn/128.2.323S. [DOI] [PubMed] [Google Scholar]
- 42.Lopez de Silanes I, Galban S, Martindale JL, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cozar-Castellano I, del Valle Machargo M, Trujillo E, et al. hIscA: a protein implicated in the biogenesis of iron-sulfur clusters. Biochim Biophys Acta. 2004;1700:179–188. doi: 10.1016/j.bbapap.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Zikova A, Panigrahi AK, Dalley RA, et al. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 46.Mofredj A, Howaizi M, Grasset D, et al. Diabetes mellitus during interferon therapy for chronic viral hepatitis. Dig Dis Sci. 2002;47:1649–1654. doi: 10.1023/a:1015852110353. [DOI] [PubMed] [Google Scholar]
- 47.Ronn SG, Billestrup N, Mandrup-Poulsen T. Diabetes and suppressors of cytokine signaling proteins. Diabetes. 2007;56:541–548. doi: 10.2337/db06-1068. [DOI] [PubMed] [Google Scholar]
- 48.Greenhalgh CJ, Rico-Bautista E, Lorentzon M, et al. SOCS2 negatively regulates growth hormone action in vitro and in vivo. J Clin Invest. 2005;115:397–406. doi: 10.1172/JCI22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tannahill GM, Elliott J, Barry AC, Hibbert L, Cacalano NA, Johnston JA. SOCS2 can enhance interleukin-2 (IL-2) and IL-3 signaling by accelerating SOCS3 degradation. Mol Cell Biol. 2005;25:9115–9126. doi: 10.1128/MCB.25.20.9115-9126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martyniak K, Masternak MM. Changes in adipose tissue cellular composition during obesity and aging as a cause of metabolic dysregulation [published online ahead of print December 9, 2016] Exp Gerontol. doi: 10.1016/j.exger.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barger PM, Kelly DP. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc Med. 2000;10:238–245. doi: 10.1016/s1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- 52.Wilson CR, Tran MK, Salazar KL, Young ME, Taegtmeyer H. Western diet, but not high fat diet, causes derangements of fatty acid metabolism and contractile dysfunction in the heart of Wistar rats. Biochem J. 2007;406:457–467. doi: 10.1042/BJ20070392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finck BN, Han X, Courtois M, et al. A critical role for PPARalpha-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation by dietary fat content. Proc Natl Acad Sci U S A. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.