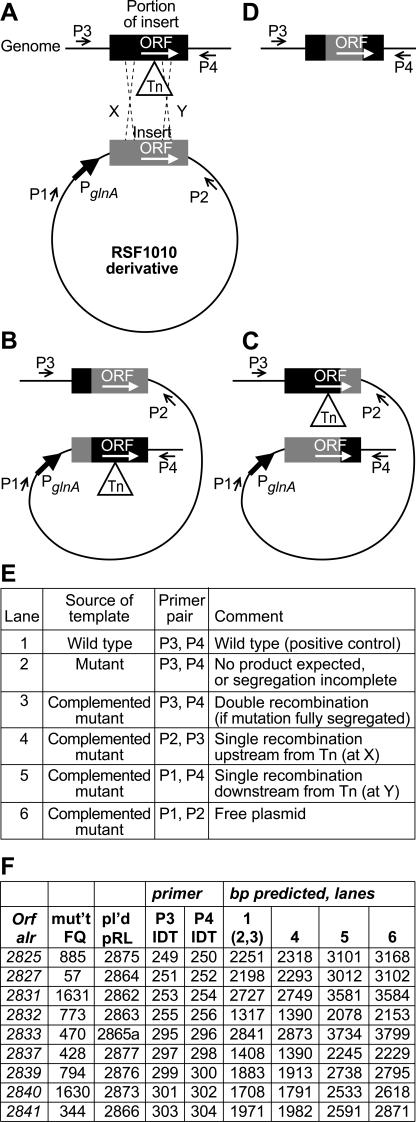
FIG.4.
Use of PCR to analyze possible recombination products in complemented mutants. (A) A plasmid (RSF1010 derivative) bears a fragment (in gray) that contains a single, intact (wild-type) copy of an ORF (shown as a white arrow) that has not recombined with the transposon (Tn)-interrupted copy of the same ORF in the genome. The ORF in the plasmid may be expressed from the glnA promoter or, perhaps, from an intervening, native promoter. PCR primers P3 and P4 are genomic sequences, up- and downstream from the cloned region, respectively, and primers P1 and P2 are vector sequences present, respectively, up- and downstream from the cloning region of the plasmid (P1 is also upstream from the glnA promoter). (B) Single, homologous recombination upstream from the transposon (at X in diagram A) would give rise to structure B, in which the uninterrupted ORF is positioned downstream from its natural upstream sequence. PCR would be expected to yield a product of predictable size with primers P2 and P3. (C) Single, homologous recombination downstream from the transposon (at Y in diagram A) would give rise to structure C, in which the uninterrupted ORF and any 3′ cotranscribed sequences would be placed under the influence of the glnA promoter and/or an intervening native promoter. PCR would be expected to yield a product of predictable size with primers P1 and P4. (D) Double, homologous recombination at both sides of the transposon would give rise to structure D, identical with that of the wild-type strain. With DNA from the wild-type strain or a double recombinant as template, PCR would be expected to yield a product of predictable size with primers P3 and P4. Such a band should be obtained with DNA from the mutant strain as template only if segregation of the mutation were incomplete, because the extension period was too short for the polymerase to traverse the transposon. If DNA from the mutant yielded no band, a band of the same size as from the wild-type strain, using DNA from the complemented mutant as template, would indicate that a double recombination event had cured the transposon from one or more copies of the genome. Lane numbers in inset tables E and F refer to lane numbers in Fig. 5, below. Inset table F shows PCR band sizes predicted (lanes 1 and 6) or predicted conditionally upon incomplete segregation (lane 2) or single (lanes 4 and 5) or double (lane 3) recombination.