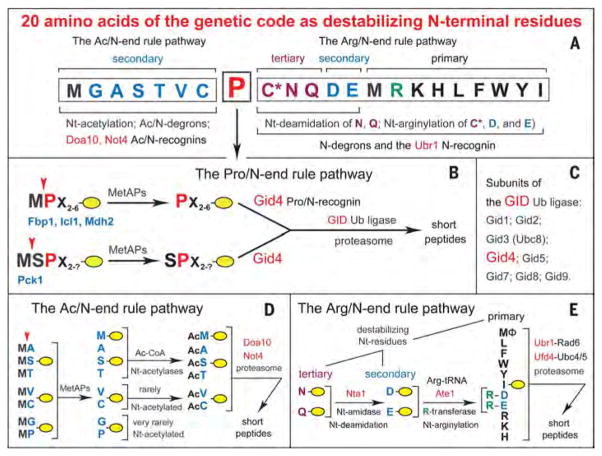
Fig. 1. Twenty amino acids of the genetic code as destabilizing N-terminal residues.
(A) N-terminal residues are indicated by single-letter abbreviations. Twenty DNA-encoded amino acids are arranged to delineate three branches of the S. cerevisiae N-end rule pathway, including its proline-specific branch, indicated by the vertical arrow. N-terminal Met is cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated Met) and by the Arg/N-end rule pathway (as the unacetylated N-terminal Met). N-terminal Cys is also cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated Cys) and by the Arg/N-end rule pathway [as an oxidized, Nt-arginylatable N-terminal Cys, denoted as Cys* and formed in multicellular eukaryotes but apparently not in S. cerevisiae (22), at least in the absence of stress]. (B) The Pro/N-end rule pathway and Gid4.We have identified this subunit of the GID Ub ligase as the Pro/N-recognin of the Pro/N-end rule pathway, whose functions include the conditional degradation of the gluconeogenic enzymes Fbp1, Icl1, Mdh2, and Pck1. A yellow oval denotes the remainder of a protein substrate. (C) Subunits of the S. cerevisiae GID Ub ligase, including Gid4. (D) The Ac/N-end rule pathway. (E) The Arg/N-end rule pathway. Amino acid abbreviations: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T,Thr; V, Val; W,Trp; Y,Tyr.