Fig. 4. Specificity of the Gid4 Pro/N-recognin.
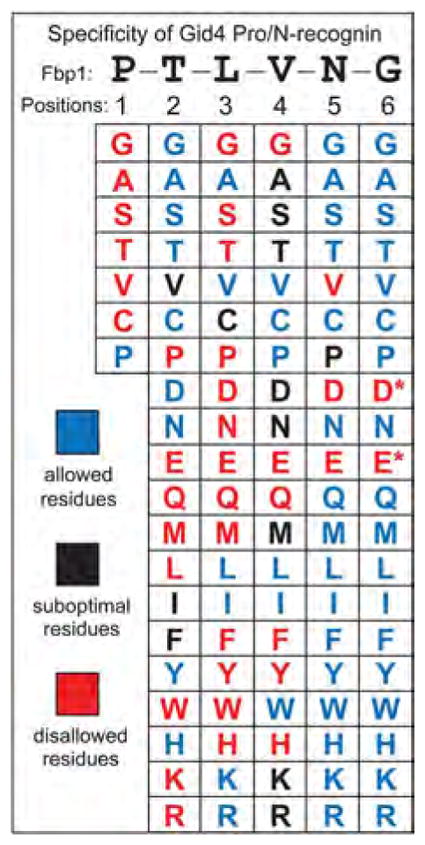
Individual residues of PTLVNG, the N-terminal sequence of wild-type P-Fbp1, were mutated to other residues while keeping residues at other positions unchanged.Two-hybrid assays were carried out to examine the binding of Gid4 to the P-X-L-V-G5-DHFR-DBD, P-T-X-V-G5-DHFR-DBD, P-T-L-X-G5-DHFR-DBD, P-T-L-V-X-G5-DHFR-DBD, and P-T-L-V-N-X-G5-DHFR-DBD fusions (X = 20 different residues) (>100 two-hybrid assays total; figs. S8 to S10). The residues are cited in this summary of the binding data as those that were “allowed (blue) (i.e., were compatible with robust binding of a fusion to Gid4), “suboptimal (black), or “disallowed (red). The latter residues abrogated the binding of Gid4 to a fusion, despite its N-terminal Pro. A residue at position 1 was varied less extensively for the reason described in the text. The Asp (D) and Glu (E) residues at position 6 are marked with asterisks to indicate that these residues were “suboptimal-like at this position, rather than abrogating the binding to Gid4. See figs. S8 to S10 for the two-hybrid data that underlie this summary.
