Fig. 5. Degradation of P-Mdh2 in cells under glucose-replete conditions.
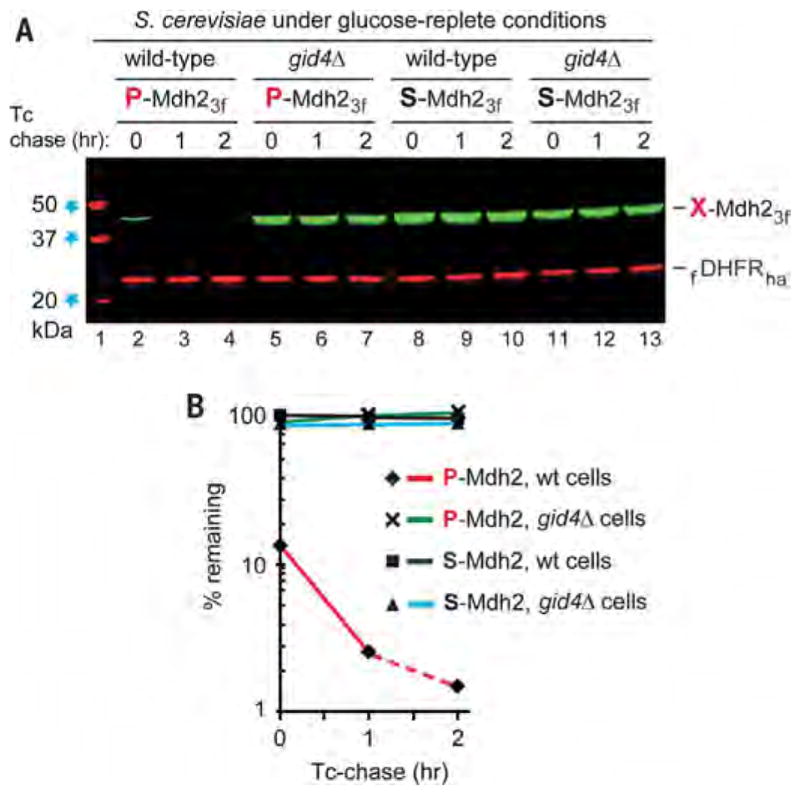
(A) Tetracycline-based, reference-based chases were performed as described in the legend to Fig. 2, at 30°C with indicated S. cerevisiae strains expressing either P-Mdh23f or S-Mdh23f, but with cells that did not undergo glucose deprivation. Lane 1: kDa markers. Lanes 2 to 4: Wild-type cells expressing P-Mdh23f. Lanes 5 to 7: Same as lanes 2 to 4 but with gid4Δ cells. Lanes 8 to 10: Same as lanes 2 to 4 but with S-Mdh23f. Lanes 11 to 13: Same as lanes 8 to 10 but with gid4Δ cells. (B) Quantification of data in (A). All chases in this study were performed at least twice and yielded results that differed by less than 10%. Note the time-zero, before-chase degradation of more than 80% of P-Mdh23f in wild-type cells (lanes 2 to 4).The bands of X-Mdh23f and fDHFRha are indicated.
