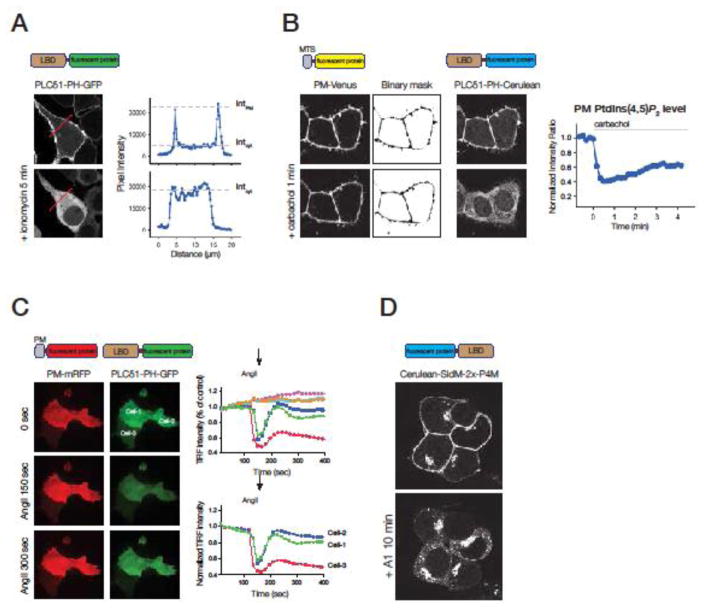
Fig 1. Methods of quantification when monitoring inositol lipids in the plasma membrane by various lipid binding domains (LBD).
(A) Various lipid binding domains (LBD) tagged with fluorescent proteins are used to follow the membrane localization of inositol lipids using confocal microscopy. Upon ionomycin treatment (causing a cytoplasmic Ca2+ increase) the plasma membrane localization of the PI(4,5)P2 specific GFP-tagged PH domain of PLCδ1 decreases in the COS-7 cells shown on the left. The translocation of the fluorescent probe can be quantified by measuring the plasma membrane and cytoplasmic pixel intensity in a line-intensity histogram (right graphs).
(B) Co-expression of a membrane-targeted fluorescent protein (MTS-fluorescent protein), such as a PM-targeted Venus (PM-Venus) allows the generation of binary masks that can be used to identify the membrane of interest during the whole measurement. The sum of the pixel intensity values of the probe defined by the binary mask gives information about the membrane bound fraction of the probe fluorescence. Images show that the localization of PM-Venus (targeted by the Lyn N-terminal 10 amino acids) is not affected by the addition of carbachol in HEK 293T cells expressing M3 muscarinic receptors. In contrast, the PH domain of PLCδ1 translocates from the PM to the cytoplasm. The process can be quantified by measuring the PM fraction of the PLCδ1-PH-Cerulean. Normalizing these values with the PM-Venus signal can be used to factor out changes of the fluorescence due to changing membrane geometry during a time-course (right graph).
(C) TIRF analysis can be used to identify the part of the PM that is attached to the tissue culture dish. Quantification of the PM-bound LBD that is proportional to the level of inositol lipids can be carried out by calculating the total pixel intensity of the cell’s footprint during a time-lapse recording. Co-expression of a PM-targeted fluorescent protein can be used to factor out intensity changes due to cell movements or changing membrane deformities. A limitation of this method that there is a signal even if the LBD is fully released from the membrane to the cytosol. This can be taken as the minimum value if one calibrates each trace at the end of experiment by a stimulus that causes complete translocation.
(D) Redistribution of an LBD-fluorescent lipid sensor upon elimination of one lipid pool. In this confocal experiment specific depletion of the PM PI4P by the treatment with a PI4KA inhibitor (A1) resulted in the disappearance of the PM-bound fluorescence, causing an increase in the Golgi- and endosome-associated signals in HEK 293T cells. PI4P was followed by expression of the Cerulean-SidM-2xP4M biosensor capable to bind PI4P both in the PM, Golgi and endosomes.