Fig 2. Quantitative measurement of inositol lipid changes using bioluminescence resonance energy transfer (BRET).
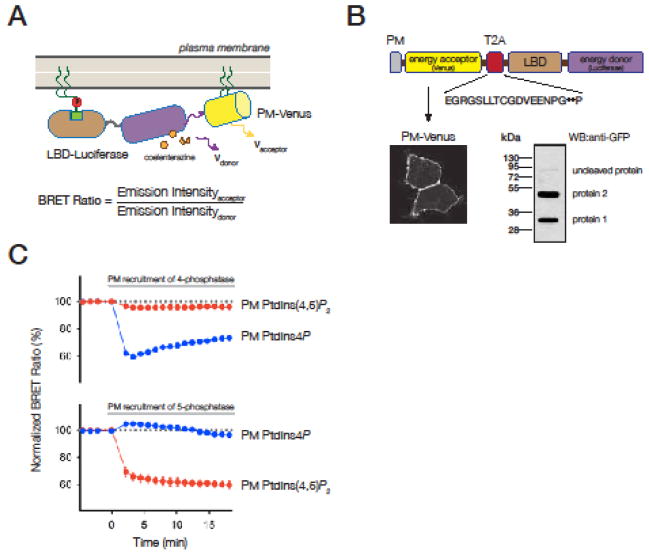
(A) Membrane localization of the LBD can be measured in cells by calculating the energy transfer between the luciferase fused to the LBD and Venus targeted to the membrane. Membrane recruitment of the probes increases, while cytoplasmic translocation of the probes decreases the BRET ratio values.
(B) Design of the BRET-based biosensors. PM-targeted Venus and the luciferase-tagged LBD (PH domain of the PLCδ1) are linked by the viral T2A protein sequence resulting in the separate expression of the two proteins. Only a small fraction of the total expressed proteins remained uncleaved as shown by the Western blot analysis of the proteins. In this experiment luciferase enzyme was replaced by Cerulean that allowed the detection of the proteins with anti GFP antibody. The confocal image shows the plasma membrane localization of Venus.
(C) HEK 293T cells were transfected with the plasmid DNA coding for components of the BRET based PI4P and PI(4,5)P2 biosensors (PM-Venus-T2A-Luciferase-2xSidM-P4M and PM-Venus-T2A-PLCδ1-PH-Luciferase, respectively). Cells were also co-transfected with plasmids encoding the rapamycin-inducible plasma membrane PI4P and PI(4,5)P2 depletion system. As shown in the graphs, the BRET biosensors can detect the changes in the respective lipid pools. Note that in case of the 5-phosphatase recruitment even the small increase of the PI4P can be detected (lower graph blue curve).