Figure 4.
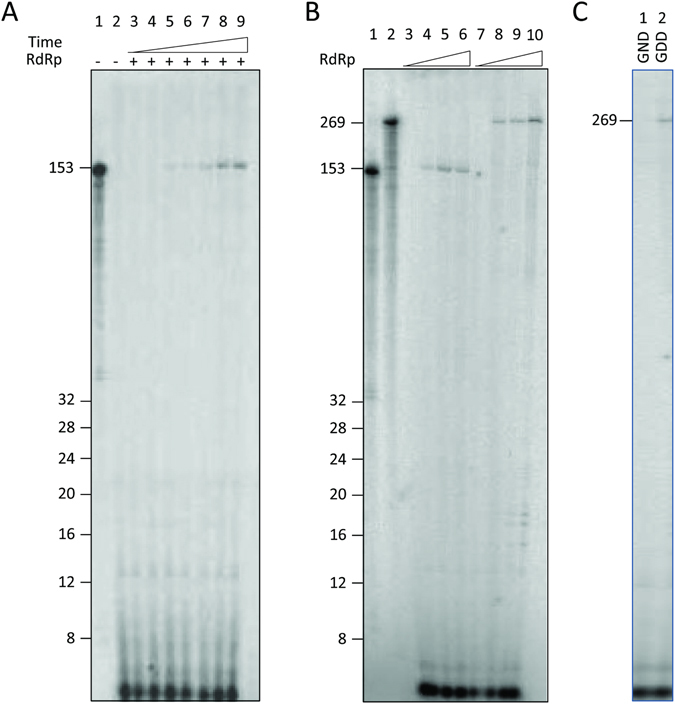
ZIKV RdRp enzymatic activities using natural templates. RNA substrates of 153 nucleotides and 269 nucleotides corresponding to the 3′-end of the (−) strand were obtained by in vitro transcription in the presence of [α32P]-UTP and subsequently used as molecular weight markers. Corresponding unlabeled RNAs were used as template to assay for the de novo RNA synthesis of the ZIKV RdRp as described in Material and Methods. (A) Time course experiment monitoring the ZIKV RdRp enzymatic activity (1.4 pmoles) using a 153–nucleotide long RNA as template. Lane 1: radiolabeled 153 nucleotides template. Lane 2: no enzyme. Lanes 3 to 9: reaction was pursued for 0, 5, 10, 15, 30, 60, 180 min, respectively. (B) Impact of the substrate length on WT RdRp activity. Lane 1 and 2: radiolabeled templates corresponding to 153 and 269 nucleotides, respectively. Lanes 3 to 6: synthesis using the 153-nucleotide long RNA substrate. Lanes 7 to 10: synthesis using the 269 nucleotides RNA substrate. Respectively 0, 0.7, 1.4, 3.5 pmoles of RdRp were used. (C) Lanes 1 and 2: Respectively, 1.4 pmoles of the GND mutant and WT RdRp (GDD) were used with the 269-nucleotide long RNA as template. Reactions were carried out for 2 h and products were separated on 12% polyacrylamide denaturing gels (7 M urea). Autoradiography was performed using an imaging plate (Fujifilm) and images were obtained with a FLA-5000 Imaging System (Fujifilm).
