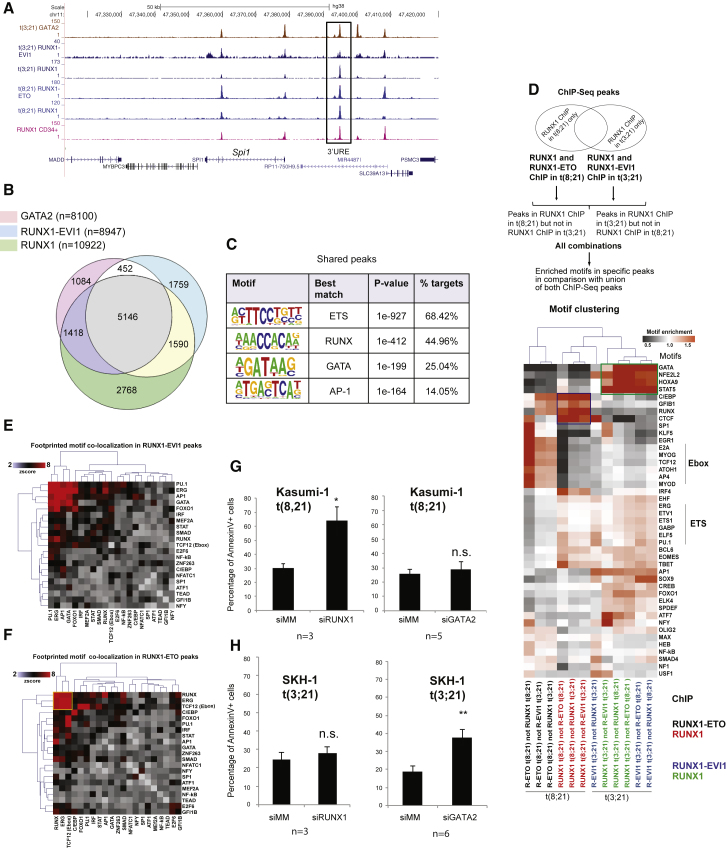
Figure 3.
RUNX1 Fusion Proteins Form Part of a Gene Regulatory Network Unique to Each Leukemia
(A) UCSC genome browser screen shot of Spi1 showing ChIP-seq data of RUNX1 and RUNX1-EVI1 from t(3;21) SKH-1 cells, RUNX1 and RUNX1-ETO from t(8;21) Kasumi-1 cells, and RUNX1 from normal CD34+ PBSCs.
(B) GATA2, RUNX1-EVI1, and RUNX1 ChIP-seq in t(3;21) SKH-1 cells. Venn diagram depicting the overlap between GATA2, RUNX1-EVI1, and RUNX1 peaks and the numbers of peaks in each group. White: overlap of GATA2- and RUNX1-EVI1-bound sites; purple: overlap of GATA2 and RUNX1 bound sites; yellow: overlap of RUNX1- and RUNX1-EVI1-bound sites; gray: number of sites bound by all three transcription factors.
(C) Transcription-factor-binding motifs enriched in the shared peaks from (B).
(D) Hierarchical clustering of enriched motifs discovered in a pairwise comparison between RUNX1 and RUNX1 fusion ChIP-seq peaks between t(3;21) and t(8;21) cells identifying unique peaks for each type of AML. Enrichment score was calculated by the level of motif enrichment in the unique peaks as compared to union of peaks in the pair of experiments. The heatmap depicts the degree of motif enrichment. Two specific sets of enriched motifs unique to each ChIP seq experiment are highlighted: the blue box highlights specifically enriched motifs in RUNX1-bound sites, the green box highlights enriched motifs specific for t(3;21) but not other peaks.
(E and F) Bootstrapping analysis of footprinted motifs at RUNX1-EVI1- or RUNX1-ETO-binding sites in patient cells. RUNX1-EVI1-binding sites from the t(3;21) SKH-1 cell line (E) and RUNX1-ETO-binding sites from the t(8;21) Kasumi-1 cell line (F) mapped onto footprints generated from DNase I data of either t(3;21) patient 2 or t(8;21) patient 1, respectively. The heatmap shows the significance of co-localizing footprinted motifs at RUNX1 fusion protein-binding sites for each AML as compared to sampling by chance alone.
(G and H) Percentage of Annexin-V-positive cells after 5 days of treatment with a control siRNA (siMM) or with siRNAs specific for RUNX1 and GATA2, respectively, in Kasumi-1 cells (G) and SKH1 cells (H). Each experiment was done at least in triplicate as indicated, and error bars represent SEM. ∗p < 0.05 and ∗∗p < 0.01 by unpaired t test. n.s. not significant.
See also Figure S3.