Figure 4.
Knockdown of RUNX1-EVI1 Results in Loss of the Stem Cell Gene Program
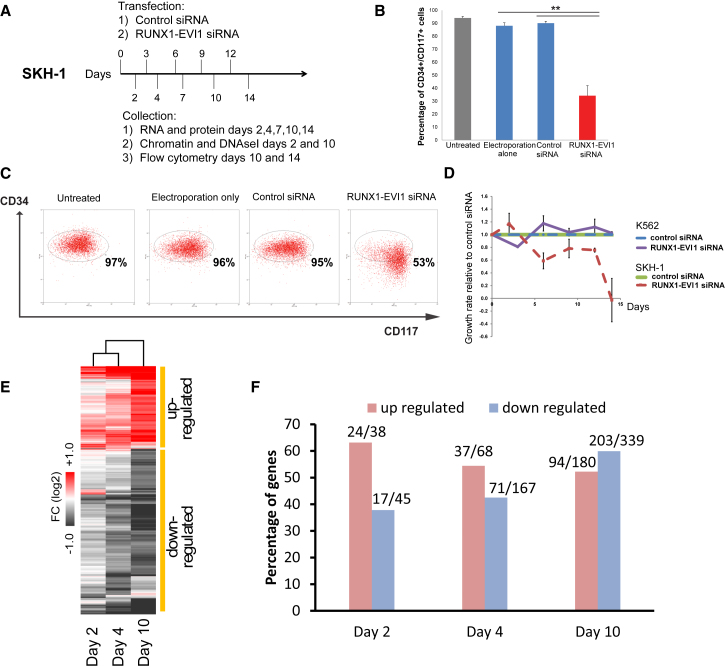
(A) Experimental scheme for the siRNA transfection.
(B and C) RUNX1-EVI1 siRNA treatment in SKH-1 cells results in reduction in CD34 surface expression. SKH-1 cells after 14 days of either RUNX1-EVI1 or control siRNA transfection were stained with CD34-PE and CD117-APC. (B) Percentage of CD34+CD117+ cells. (C) Representative flow cytometry plot. Mean of six independent experiments is shown. Error bars represent SEM. ∗p < 0.05 by paired t test.
(D) Growth rates of SKH-1 (dashed lines) and K562 cells (solid lines) treated with RUNX1-EVI1 siRNA relative to treatment with control siRNA. The graph shows mean and SEM values from at least three independent experiments.
(E) Hierarchical clustering of gene expression changes as determined by RNA-seq at different time points of treatment. Unsupervised clustering of expression values of genes changing expression 1.5-fold after RUNX1-EVI1 siRNA transfection as compared to control siRNA. Average of two independent replicates. The heatmap color is related to the degree of differential expression (fold change [FC]) between RUNX1-EVI1 siRNA and control siRNA treatment.
(F) Percentage and number, respectively (on top of bars), of differentially expressed genes as measured by RNA-seq that are RUNX1-EVI1 ChIP-seq targets. Differentially expressed genes are those with an at least 1.5-fold change in gene expression between RUNX1-EVI1 siRNA as compared to control siRNA treatment.
See also Figure S4.