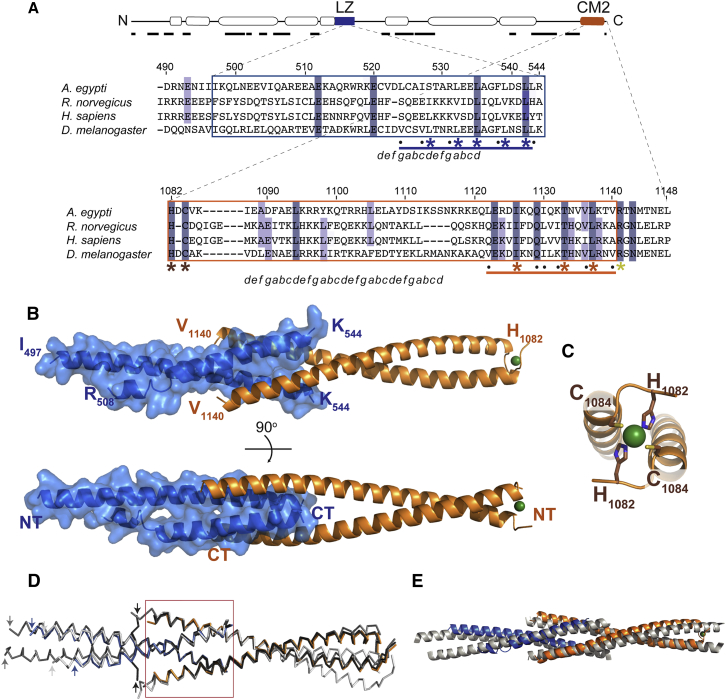
Figure 2.
Crystal Structure of the LZ:CM2 Complex
(A) Schematic illustration of Drosophila Cnn highlighting predicted coiled-coil regions (bubbles; predicted by COILS [Lupas et al., 1991]), predicted disordered regions (black lines; predicted by XtalPred-RF [Slabinski et al., 2007]), and the LZ (blue) and CM2 (orange) domains. Expanded regions show multiple sequence alignments (MSAs) of the regions used for crystallization (see Figure S1A for a more comprehensive MSA of the CM2 domain); boxed regions indicate residues visible in the crystal structures. Bars indicate the interaction interface with dots or asterisks over the bars highlighting residues buried in the interface. Asterisks highlight residues subjected to mutational analysis. Residues identified by SOCKET (Walshaw and Woolfson, 2001) as belonging to a canonical coiled coil in the structure are annotated beneath the sequence with a–g lettering.
(B) Side and top views of the LZ (blue):CM2 (orange) complex, shown in cartoon representation; a space-filling model of LZ is overlaid with a reduced opacity. The coordinating Zn2+ ion is shown as a green sphere. The N terminus (NT) and C terminus (CT) of each protein are indicated.
(C) Close up view of the N-terminal region of CM2 highlighting the coordination of the Zn2+ ion.
(D) Ribbon diagram of three different LZ:CM2 domain crystal structures (shades of gray) overlaid on the original LZ:CM2 structure (blue and orange) shown in (B); the N termini of the different LZ constructs are indicated by arrows. The core of the LZ:CM2 interaction interface is similar in all of the structures (red box), but the surrounding helical regions exhibit considerable variation.
(E) An overlay of the LZ:CM2 structure (blue:orange) and the Homer1:Homer1 tetramer (gray) (PDB: 3CVE). See also Figures S1 and S2.