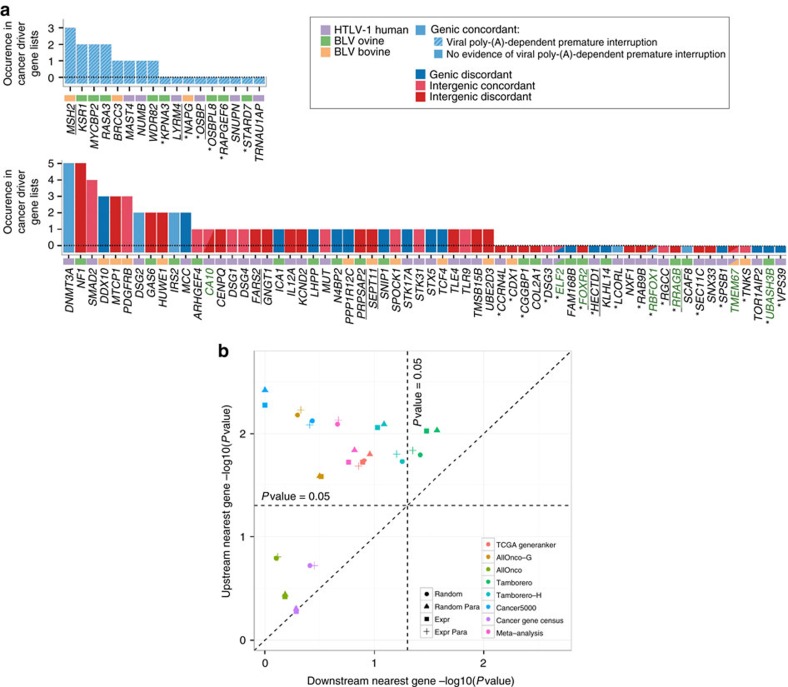
Figure 4. HTLV-1/BLV interacting host genes are connected to cancer.
(a) Transcript-interacting host genes were ranked according to their occurrence in seven cancer driver lists used for enrichment (Table 1 and Supplementary Table 2). Top panel: viral poly-(A) truncated genes disrupted by genic-concordant proviruses. Bottom panel: all genes interacting with 3′AS-dependent transcripts. Cancer drivers are equally represented between provirus types (genic, intergenic) and across species. Underlined genes: genes for which transcriptional patterns in tumours are shown in Figs 1 and 3 and Supplementary Fig. 5. Recurrent genes between tumours: green symbols. *Genes absent from cancer driver lists for which literature screen supported connection to cancer. This list comprises FOXR2, RRAGB, ELF2 and SPSB1 (refs 40, 42, 66, 68), genes that exhibit undeniable oncogenic properties. UBASH3B (BLV, sheep): identified as the target gene of HTLV-1 integration in one of the ATLs analysed in a recent WGS study11. The remaining protein-coding genes and interacting non-coding RNAs not previously reported in cancer (Supplementary Data 2 and Supplementary Table 4, antisense transcript-interacting lncRNAs) represent a potential resource of novel candidate cancer drivers of both the coding and noncoding class of genes. (b) Host genes upstream (y-axis) of proviral integration sites in ATLs and B-cell tumours (92 sites) show significant enrichment in cancer drivers in contrast to the corresponding downstream host genes (x-axis), supporting antisense-dependent cancer driver perturbation by HTLV-1/BLV proviruses. The direct target genes of genic proviruses were excluded from the analysis. The significance of the enrichment was computed for seven publicly available cancer driver lists and for all list combined by means of a meta-analysis (Supplementary Data 3 and Supplementary Table 2). Observed scores were compared to simulated scores obtained from N=100,000 size-matched random or expression-matched gene sets, including information about paralogs (Random Para, Expr Para) or not (Random, Expr) (Supplementary Fig. 2). Symbol code: simulated gene sets, colour code: cancer driver lists.