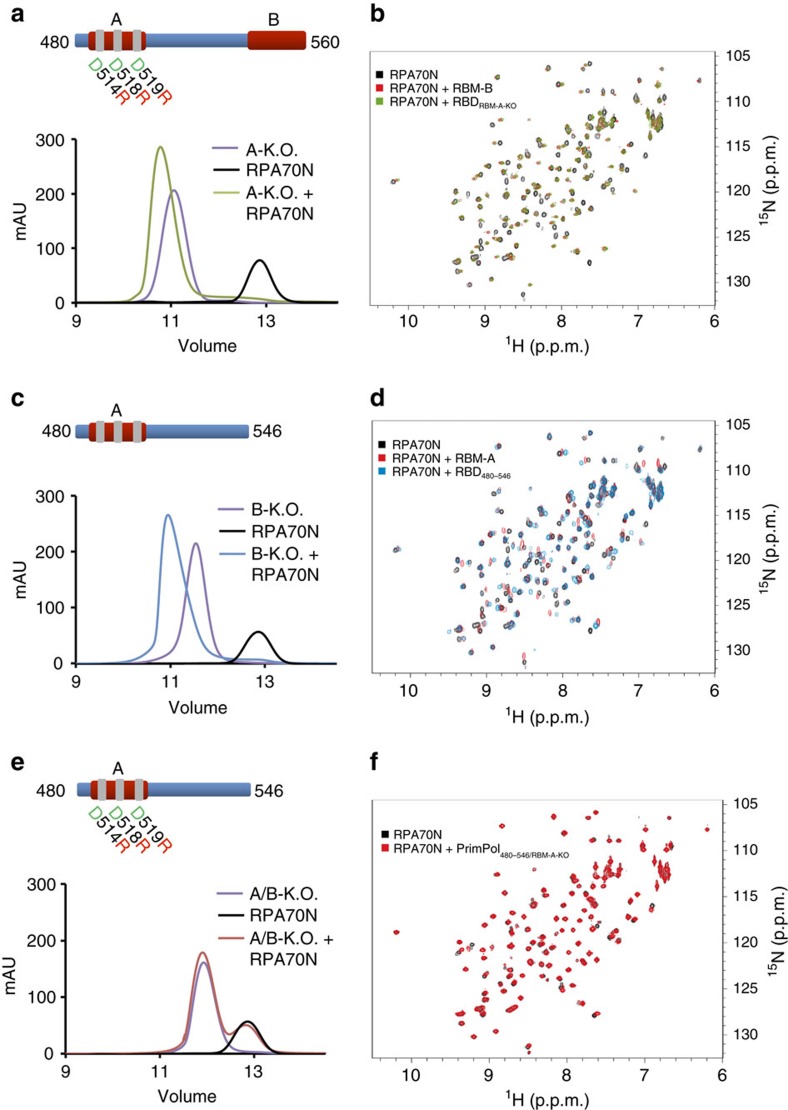
Figure 3. RPA70N dynamically interacts with both RBM-A and RBM-B.
(a) Mutation of RBM-A does not abolish binding of PrimPol's RBD to RPA70N. Chromatographs showing the retention volumes of RBDA-KO (purple), RPA70N (black) and RBDA-KO with RPA70N in a 1:1 ratio (green). (b) 15N-1H HSQC spectra showing RPA70N alone (black), in the presence of twofold molar excess of either RBDA-KO (green) or RBM-B peptide (residues 542–560) (red). The perturbations observed for RBDA-KO are similar to those induced by the RBM-B peptide. (c) Truncation of RBM-B does not prevent binding of PrimPol's RBD to RPA70N. Chromatographs showing the retention volumes of RBDB-KO (purple), RPA70N (black) and RBDB-KO with RPA70N in a 1:1 ratio (blue). (d) 15N-1H HSQC spectra showing RPA70N alone (black) or in the presence of twofold molar excess of RBDB-KO (blue) or RBM-A peptide (residues 510–528) (red). The perturbations observed for RBDB-KO are similar to those induced by the RBM-A peptide. (e) Mutation of both RBM-A and RBM-B abolishes the binding of PrimPol's RBD to RPA70N. Chromatographs showing the retention volumes of RBDA/B-KO (purple), RPA70N (black) and RBDA/B-KO with RPA70N in a 1:1 ratio (red). (f) 15N-1H HSQC spectra showing RPA70N alone (black) or in the presence of twofold molar excess of RBDA/B-KO (red). The near identity of the two spectra indicates there is no interaction.