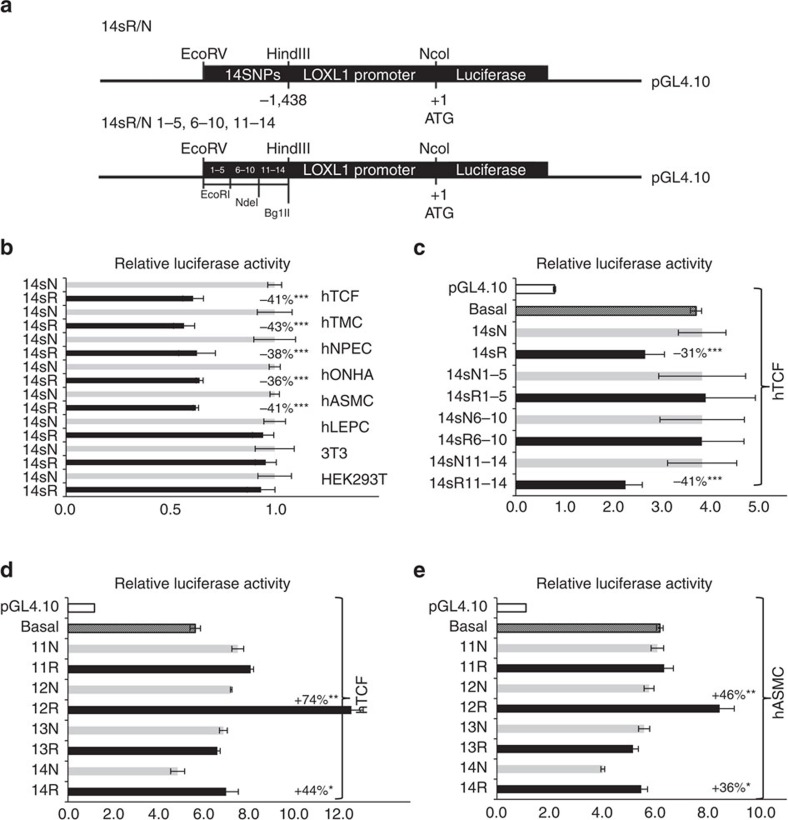
Figure 5. Effects of risk variants on LOXL1 promoter transcriptional activity in vitro.
(a) Reporter constructs used in transfection experiments outlining the 14 S/R constructs comprising all 14 SNPs flanked by 51 bp of genomic DNA sequences each (top) and three deletion constructs comprising SNPs 1–5, SNPs 6–10 and SNPs 11–14, respectively (bottom). (b) Dual luciferase reporter assays demonstrating regulatory activity of 14 SNP risk (14sR) and non-risk (14sN) haplotypes in human Tenon‘s capsule fibroblasts (hTCF), trabecular meshwork cells (hTMC), nonpigmented ciliary epithelial cells (hNPEC), optic nerve head astrocytes (hONHA), aortic smooth muscle cells (hASMC), limbal epithelial cells (hLEPC), 3T3 fibroblasts and HEK293T cells. Results are expressed as the ratio of Firefly luciferase to Renilla luciferase; the transcriptional activity of the non-risk sequence was set at 100%. (c) Regulatory activity of three deletion constructs (SNPs 1–5, 6–10 and 11–14) in hTCF compared with the basal LOXL1 promoter activity; the transcriptional activity of the empty pGL4.10 vector was set at 100%. (d,e) Activity assays using reporter plasmids containing each of the four risk or non-risk alleles of individual SNPs 11, 12, 13 and 14 compared with the basal LOXL1 promoter activity in hTCF (d) and hASMC (e); the transcriptional activity of the empty pGL4.10 vector was set at 100% (data represent mean values±s.d. of five independent experiments; *P<0.05; **P<0.005; ***P<0.0001; unpaired two-tailed Student‘s t-test).