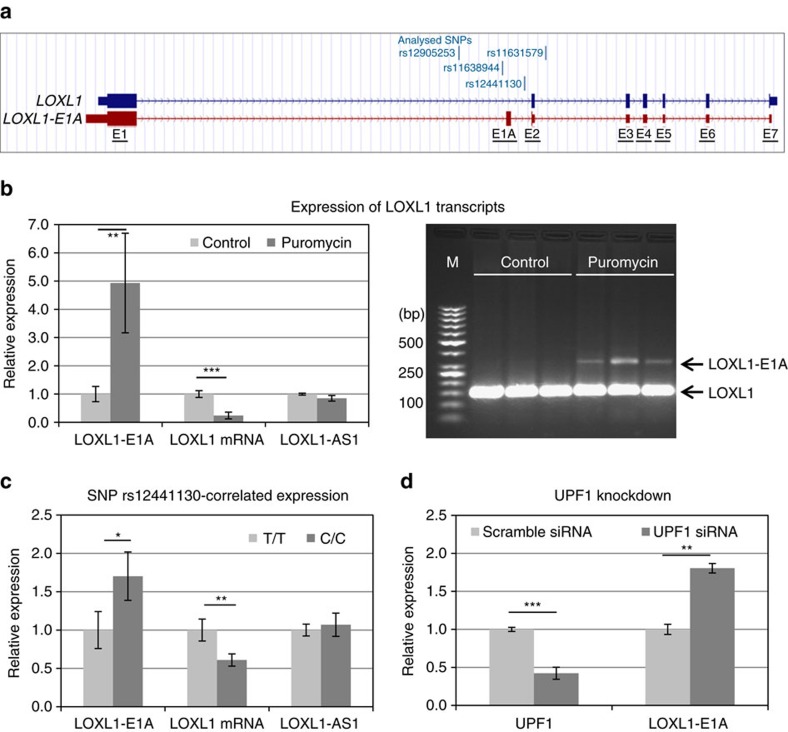
Figure 7. Effects of risk variants on LOXL1 alternative splicing.
(a) The two LOXL1 transcripts (LOXL1, LOXL1-E1A) seen in the alignment with position of SNPs 11–14; exons are indicated by numbers (E1–E7), the additional exon included in the alternative transcript is indicated as E1A; the location of stop codons is marked by vertical lines. (b) Expression levels of transcripts LOXL1-E1A, LOXL1 and LOXL1-AS1 in hTCF cell lines (n=16) without (control) or after treatment with puromycin (200 ng ml−1 for 10 h) using real-time PCR technology (left); hTCF were either homozygous for the risk (n=8) or non-risk (n=8) alleles for SNPs 1–14. PCR from LOXL1 and LOXL1-E1A transcripts without and after puromycin treatment of hTCF (n=3) using primer pairs spanning exon 1A (right; lane M: DNA marker). (c) SNP rs12441130-correlated expression levels of transcripts LOXL1-E1A, LOXL1 and LOXL1-AS1 in hTCF cell lines homozygous for the risk alleles (C/C) (n=8) or non-risk alleles (T/T) (n=8) using real-time PCR technology. (d) Real-time PCR analysis of UPF1 and LOXL1-E1A in hTCF cell lines (n=4) transfected with UPF1-specific siRNA or scrambled control siRNA; expression levels were normalized relative to GAPDH (data represent mean values±s.d.; *P<0.05; **P<0.001; ***P<0.0001; unpaired two-tailed Student‘s t-test).