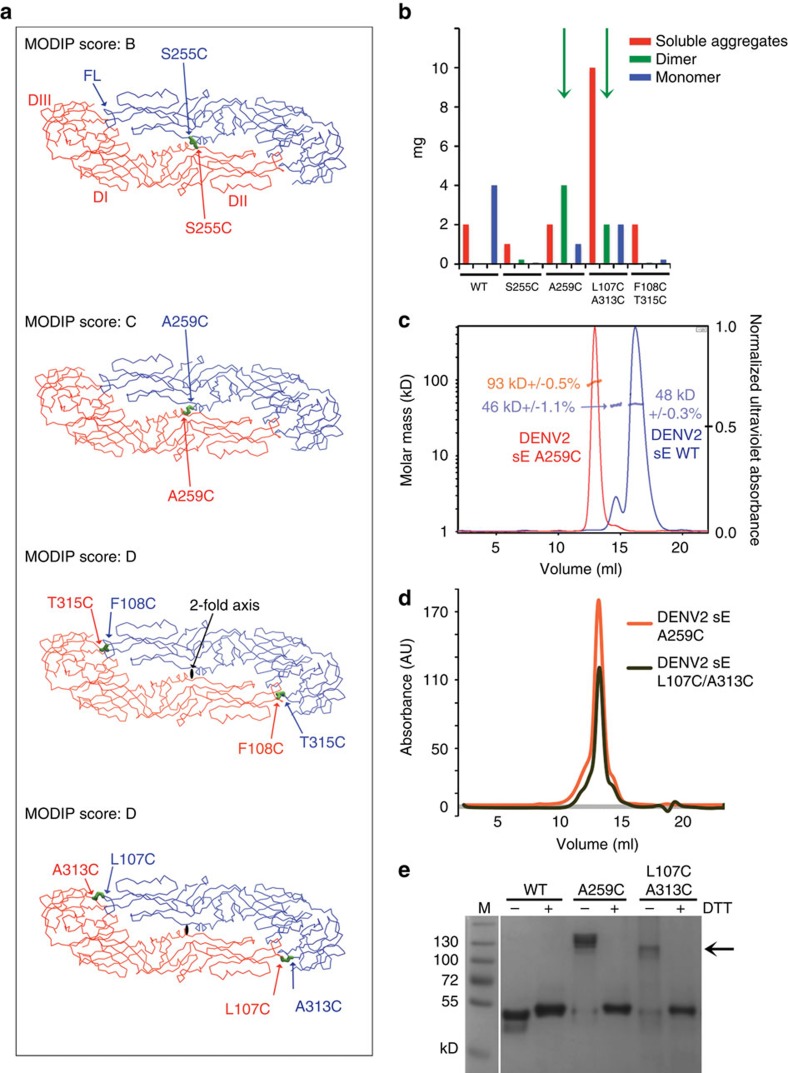
Figure 3. Engineering covalently linked E-dimers.
(a) Localization of the residues identified by MODIP susceptible to form inter-chain disulfide bonds upon mutation to cysteine. The sE dimer is coloured by subunit, with the MODIP residue pairs indicated in the corresponding colours. A disulfide bond is modelled and is shown as green sticks. The MODIP score, indicated for each residue pair, is a measure of favourability of the geometry of the selected amino acids for disulfide bond formation where A is best and D is worst. (b) Histogram showing the approximate yields in mg per litre of S2 cell culture of DENV2 FGA02 sE protein eluting as monomer, dimer and aggregates separated by SEC for wild type and for the four cysteine mutants presented in a. The yields of covalent dimers are shown in green bars (highlighted with green arrows when sufficient yields for further studies were obtained). (c) MALS analysis of DENV2 A259C sE (red trace). The fractions eluting as dimer in a first step of SEC (which eliminated monomers and aggregates) was re-run by SEC and then superposed to the elution profile of DENV2 WT sE (blue trace). The ultraviolet absorbance was normalized such that the highest peak of each run is set to 1 (y axis on the right). The molecular weight determined by MALS is indicated, corresponding to the y axis on the left. (d) SEC elution profile of L107C/A313C sE superposed to that of A259C sE, showing that the peaks are at the same elution volume, which corresponds to a dimer characterized by MALS in b. As in c the peaks corresponding to monomer and aggregates were eliminated in an initial SEC run. (e) Coomassie stained SDS-PAGE run of sE WT and sE mutants of DENV2 (in the absence (−) or presence (+) of reducing agent DTT). The black arrow indicates the bands of the disulfide stabilized sE dimer.