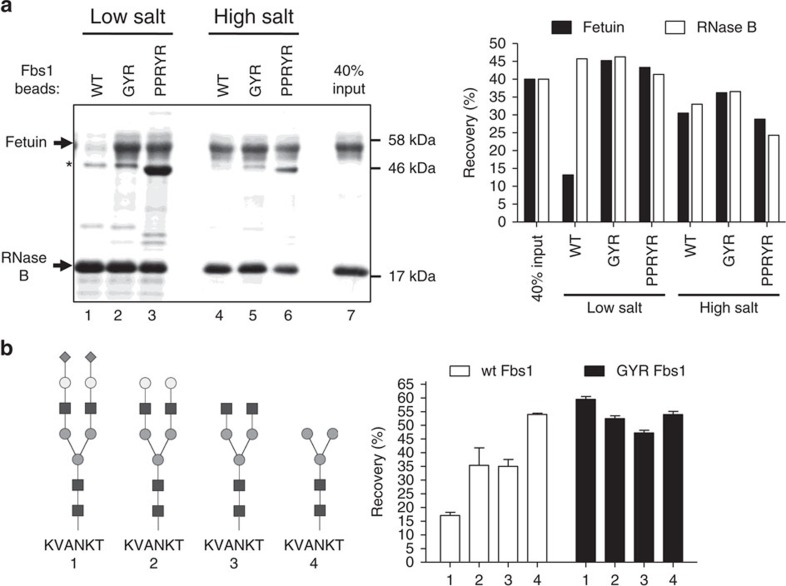
Figure 4. Fbs1 GYR and PPRYR variants display reduced binding bias between high-mannose and complex N-glycans.
(a) Comparison of N-glycoprotein pulldown by wt Fbs1, Fbs1 GYR and PPRYR variant proteins. A mixture of denatured RNase B and fetuin was subjected to an Fbs1 pulldown assay with wt, GYR and PPRYR Fbs1 beads in low salt (50 mM ammonium acetate, pH7.5) and high salt (2M ammonium acetate, pH7.5). All three Fbs1 bead types were conjugated with the same amount of the respective Fbs1 protein (Supplementary Fig. 1). Left panel is the SDS–PAGE gel showing the bound (Lanes 1–6) and input ratio (Lane 7) of RNase B and fetuin. An asterisk denotes the SNAP-Fbs1 protein leaching from the Fbs1 beads. Right panel shows the recovery percentage (bound protein amount/input protein amount) of each substrate glycoprotein using the different conditions. A representative SDS–PAGE gel is shown from two experiments. (b) Fbs1 GYR variant binding to a diverse set of N-glycopeptides is substantially unbiased. The experiment in Fig. 1d was repeated using Fbs1 GYR beads. The data shown in Fig. 1d are presented in this figure to facilitate the comparison between wt Fbs1 and Fbs1 GYR. N-glycans of SGP-TMR (1) were trimmed with different combinations of exoglycosidases to produce asialo-SGP-TMR (2), SGP-TMR without sialic acids and galactose (3) and SGP-TMR without sialic acids, galactose and GlcNAc (4). Identities of the trimmed SGP-TMR derivatives were confirmed by LC-MS. The trimmed glycopeptides were then added to binding assays with wt Fbs1 or Fbs1 GYR beads in 50 mM ammonium acetate pH7.5. The relative binding affinity to wt Fbs1 or Fbs1 GYR is reported as the recovery percentage (TMR fluorescence on beads/input TMR fluorescence). For simplicity, TMR is not shown in the N-glycopeptide structures (1–4). Results represent the mean±s.e.m. of three replicates.