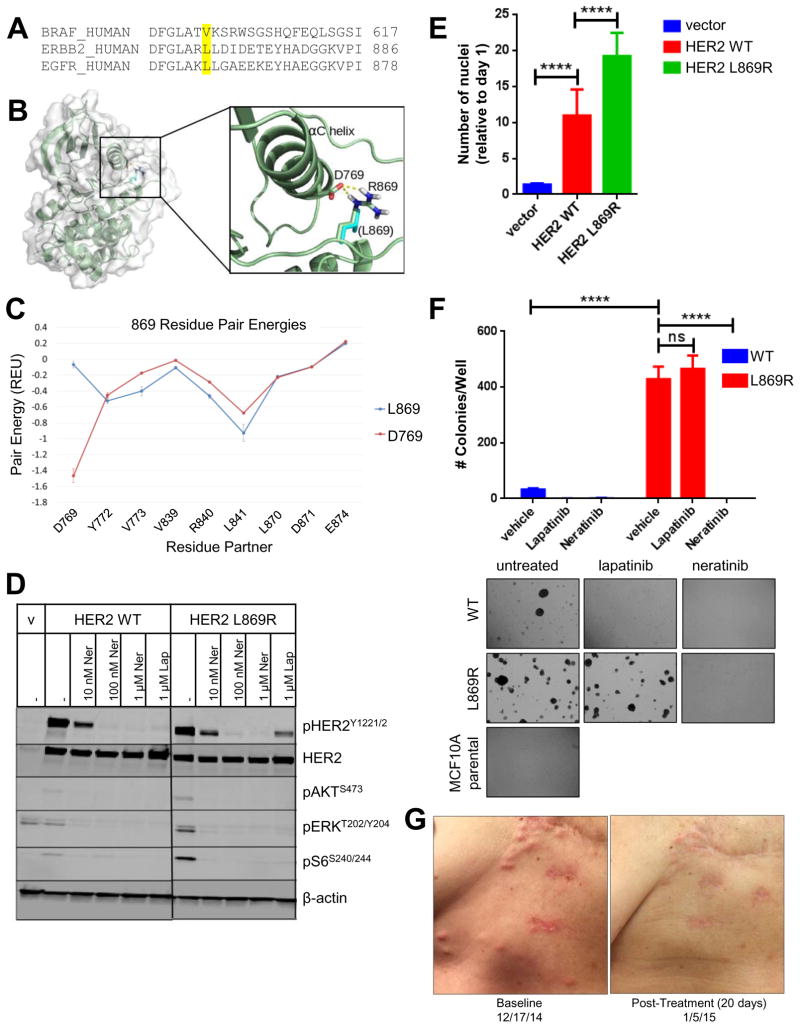
Fig. 1. HER2L869R exhibits a gain-of-function phenotype that is blocked by neratinib.
(A) The amino acid sequences of human BRAF, ERBB2, and EGFR were aligned using Clustal Omega. BRAF V600, ERBB2 L869, and EGFR L861 residues are highlighted in yellow. (B) The structure of HER2L869R was modeled. The mutation from Leucine (cyan) to Arginine (highlighted in blue) permits favorable charge interaction (dashed yellow lines) with Asp769. (C) Residue pair energies involving residue 869 reveal the addition of a strong attractive (negative) interaction at Asp769 in the HER2L869R model. (D) MCF10A cells stably expressing HER2WT or HER2L869R were treated with vehicle (DMSO), 0.01–1.0 μM neratinib, or 1 μM lapatinib for 4 h in serum-free media. Cell lysates were probed with the indicated antibodies. Scans are all from the same gel/film; the vertical black line indicates an irrelevant lane that was removed from the figure for clarity. (E) Stably transduced MCF10A cells were seeded in 96-well plates in MCF10A starvation media (1% charcoal-stripped serum, no EGF). After 7 days, nuclei were stained with Hoechst and scored using the ImageXpress system. Data points represent the average ± standard deviation (SD) of 4 replicate wells (****, p<0.0001, ANOVA followed by Tukey’s multiple comparisons test). (F) Stably transduced MCF10A cells were plated in 3D Matrigel in presence of the indicated drugs (100 nM). Colonies were grown in media containing 5% charcoal-stripped serum without EGF and insulin. After ~2 weeks, colonies were stained with MTT and counted using the Gelcount system. Data represent the average ± SD of 3 replicates. Representative fields (10X objective) of wells are shown below (****p<0.0001, ANOVA followed by Tukey’s multiple comparisons test). (G) Chest wall skin metastases of patient with invasive lobular breast cancer harboring HER2L869R at baseline and 20 days after starting treatment with neratinib.