Figure 2.
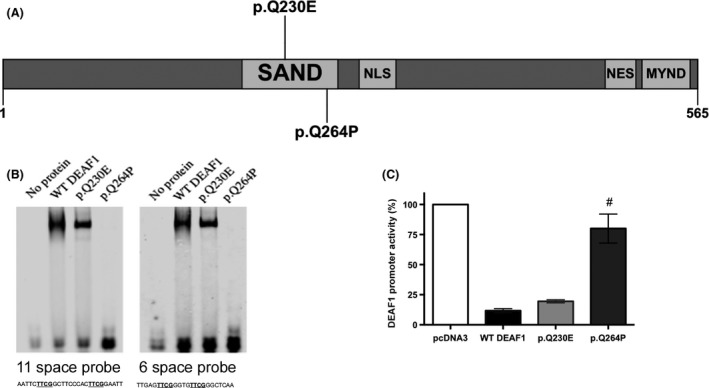
in vitro functional studies of the DEAF1 variant (p.Q230E) (A) A schematic of DEAF1 (565 domains) and the locations of a known pathogenic DEAF1 variant (p.Q264P) in gray and our proband's DEAF1 variant (p.Q230E) in yellow. (B) EMSA of WT and altered FLAG‐tagged DEAF1 recombinant proteins isolated from HEK293T cells. Fluorescently labeled DNA ligands with a spacing of 11 or 6 nucleotides between the CG dinucleotides were examined. The positive control protein, p.Q264P proteins, lacked DNA binding, whereas the p.Q230E protein showed similar binding to the WT DEAF1. (C) DEAF1 promoter activity after transfection with either WT or altered FLAG‐tagged DEAF1. The p.Q230E substitution had no effect on transcriptional repression. The p.Q264P served as a positive control of a known deleterious DEAF1 variant. Each bar represents the mean ± SEM of the normalized luciferase activity of three independent experiments when the activity of pcDNA3 (DEAF1 promoter alone) was set to 100%. One‐way ANOVA with both Dunnett's multiple comparisons and selected Bonferroni post‐test of WT DEAF1 versus each mutant, # P < 0.001.
