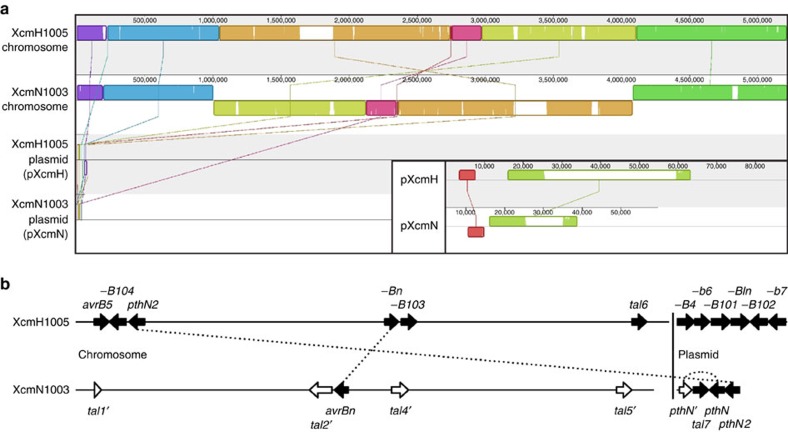
Figure 1. Comparison of whole genomes and tal genes of XcmH1005 and XcmN1003.
(a) Alignment of the XcmH1005 and XcmN1003 genomes, generated using progressiveMAUVE with default parameters68. Each genome comprises a single circular chromosome and a single circular plasmid, shown linearized. Coloured, rounded rectangles represent locally collinear blocks (LCB), regions of homology without rearrangement across the aligned sequences, connected by matching coloured diagonal lines. The orientations of the LCB, forward or reverse, are indicated by their position above or below the line, respectively. The height of a column within a block reflects the average similarity relative to the other aligned sequence(s) there (see http://darlinglab.org/mauve/user-guide/viewer.html for details). Inset (bottom right) shows the alignment of the plasmids only, at a larger scale. Horizontal axes show sequence coordinates (bp). MAUVE backbone files giving the exact coordinates of all LCB are provided as Supplementary Data 1 (all molecules aligned) and Supplementary Table 1 (plasmids only). (b) The tal genes of XcmH1005 and XcmN1003. The genes are represented as block arrows at their relative positions in the chromosome or plasmid (horizontal lines). The arrows are magnified relative to the rest of the genome, but intergenic regions and arrow sizes relative to each other are to scale. Dashed lines connecting two arrows indicate identical encoded RVD sequences. Gene names follow the scheme of ref. 41, except for the indicated avr and pth genes, named previously10,38,39,40. For label clarity, hypens replace ‘avr'. An apostrophe following the gene name (white arrows) indicates that the coding sequence is terminated early due to a frameshift mutation or other coding sequence disruption. Details, including RVD sequences and AnnoTALE designations53, are given in Supplementary Table 2.