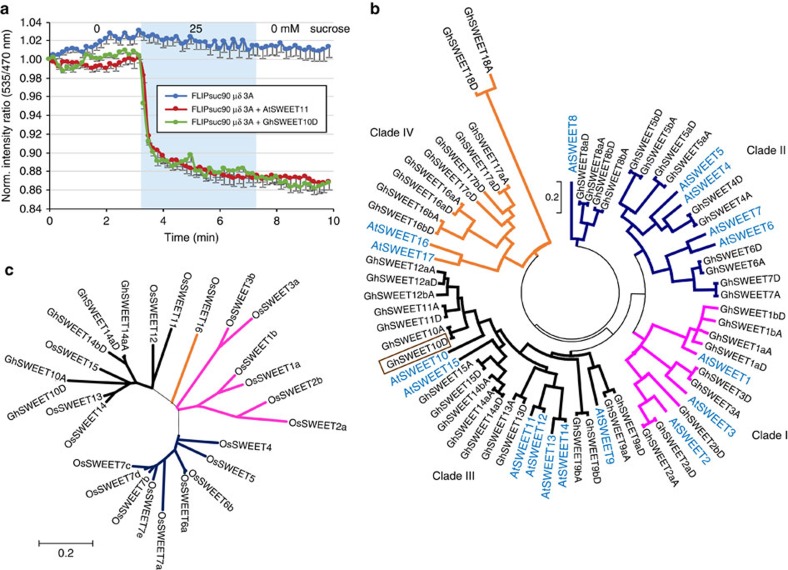
Figure 6. GhSWEET10D encodes a functional clade III sucrose transporter.
(a) Detection of GhSWEET10D transporter activity in HEK293T cells using the sucrose sensor FLIPsuc90μδ3A. Sucrose transporter activity was assayed by co-expressing GhSWEET10D with the cytosolic FRET sucrose sensor FLIPsuc90μδ3A in HEK293T cells. Blue circles correspond to cells expressing the sensor alone; red circles and green circles correspond to cells co-expressing the sensor with AtSWEET11 or GhSWEET10D, respectively. The cyan block indicates duration of perfusion with 25 mM sucrose. Accumulation of sucrose is reported as a negative ratio change (mean−s.e.m.; n>10). Experiments were repeated four times with similar results. (b) Phylogenetic analysis of GhSWEET proteins from G. hirsutum. AtSWEET proteins from Arabidopsis thaliana were included to represent different clades of SWEET superfamily. The phylogenetic tree was made using the neighbour-joining method in MEGA version 6.06. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site69. GhSWEET10D, which was used as the query, is highlighted with a brown box. (c) Phylogenetic analysis of GhSWEET10, GhSWEET14a, GhSWEET14b and the OsSWEET proteins from Oryza sativa. The phylogenetic tree was made as described in b. The clades of the SWEET family were colour coded as follows: clade I=pink, clade II=blue, clade III=black and clade IV=orange.