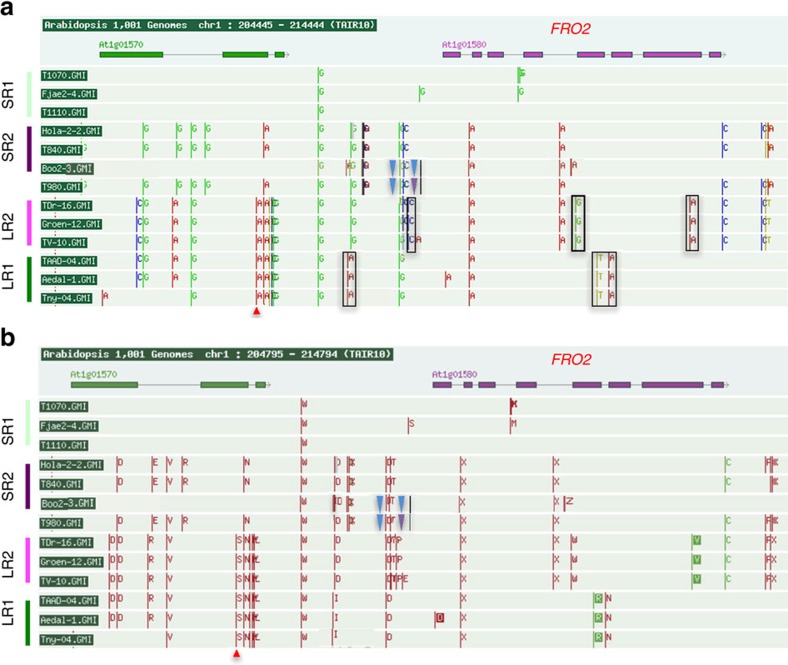
Figure 5. SNP polymorphisms around the FRO2 locus in extreme accessions of distinct FRO2 haplogroups.
SNP polymorphisms (a) and amino acid changes (b) surrounding the FRO2 locus in three representative accessions from the four main haplotypes (faint green (SR1) and dark pink (SR2) haplotypes with short root phenotype and faint pink (LR2) and dark green (LR1) with long root phenotype). Red arrow: location of the significant GWA SNP. Black boxes: Most likely candidate SNPs underlying the allelic effects. SNP polymorphism in FRO2 regulatory and coding region is constructed based on Sanger sequencing data. SNP polymorphisms in Boo2-3, T980, and TNY 04 accessions are modified based on Sanger sequencing data (Black line: 1 bp deletion, blue triangle: 2 bp insertion, purple triangle: 3 bp insertion). Only genomes that were available on the SALK 1,001 genomes browser (http://signal.salk.edu/atg1001/3.0/gebrowser.php) as of September 2016 were considered.