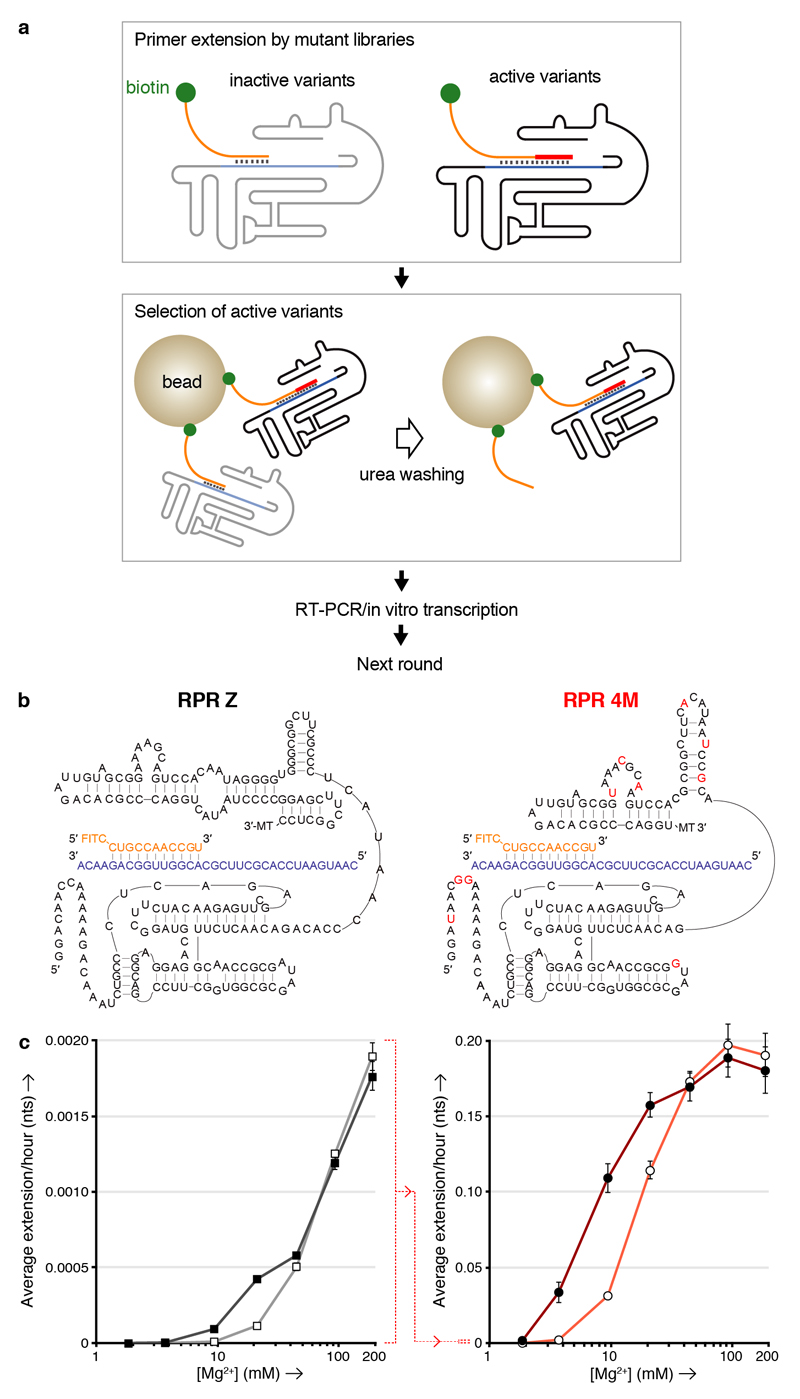
Figure 3. K10 and [Mg2+] dependence of RPR activity by Z and the evolved 4M.
a, Outline of the TST selection scheme, showing how in cis-primer extension by a ribozyme prevents ribozyme loss during a denaturing wash on beads, allowing it to be recovered and amplified. b, Putative secondary structures of the Z and 4M ribozymes surrounding the primer (orange)/template (purple) duplex, showing 4M selected mutations in red (3’ MT = tail sequence from selection construct). c, Average primer extension per hour on template TI by the Z (left) or 4M (right) RPRs in 5 d (left) or 4 h (right) reactions at varying [Mg2+], with 6 μM K10 (filled symbols) or without it (open symbols) (error bars represent S. D., N = 3; note the change in scale indicated between the two panels (dotted red line)).