Figure 5. ZRF1 is present in the nucleolus and causes relocalization of chromatin.
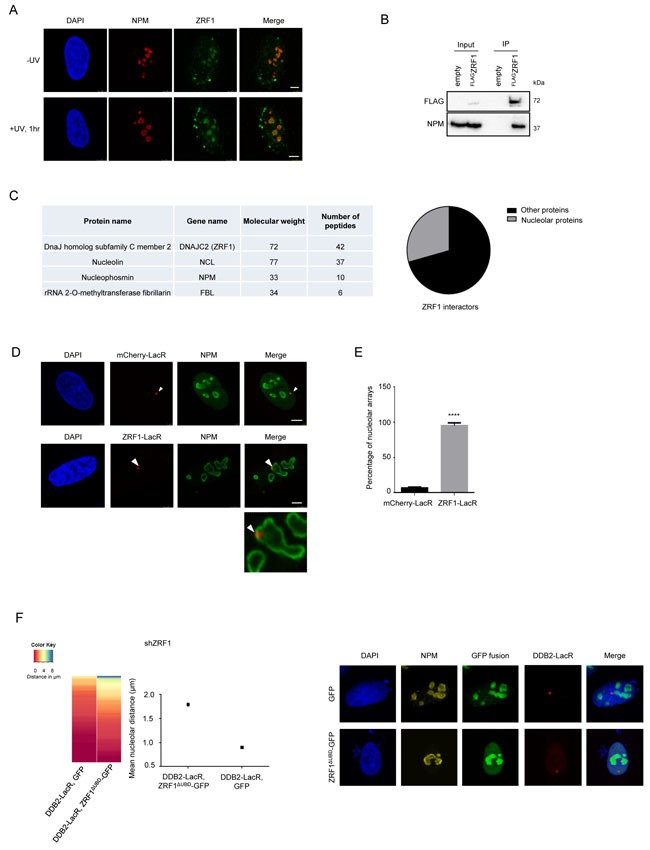
A. ZRF1 localizes to nucleoli. Immunofluorescence images showing intra-nuclear distribution of ZRF1 in pre-extracted control cells and cells exposed to UV irradiation. Nucleoli are marked by Nucleophosmin (NPM). Pre-extraction washes off unbound nuclear and cytoplasmic proteins and enables visualization of chromatin-associated proteins. Scale bar: 5μm. B. ZRF1 interacts with NPM. Purifications from HEK293T cells transfected with either empty vector or FLAGZRF1, show specific interaction of NPM with ZRF1 C. ZRF1 interacts with major components of the nucleolus. FLAGZRF1, along with its interactors, was purified from HEK293T cells and the purified material was subjected to mass spectrometry. Multiple components of the nucleolus were found to interact with ZRF1. (Left panel) the table shows peptide numbers for selected proteins in the FLAG purification. (Right panel) 30% of all interacting proteins were found to be nucleolar, as assessed by comparison with the nucleolar protein database NOPdb (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1347367/#!po=8.62069). D. Tethering of ZRF1 to a LacO array leads to repositioning of the array. Representative images of mCherry-LacR-ZRF1 or mCherry-LacR tethered LacO arrays. Nucleoli are marked by Nucleophosmin (NPM). Scale bar -5μm E. Quantification of nucleolar arrays (distance from nucleolus = 0μm) in control (mCherry-LacR) and ZRF1 (ZRF1-LacR) tethered arrays. 90-100 cells were counted per experiment. The graph shows the mean ±SD. (n = 3). Significance was determined by an unpaired t-test. F. Overexpression of ZRF1ΔUBD-GFP abolishes repositioning of a DDB2-LacR tethered array. (Left panel) Heat map showing distribution of the nucleolar distance of the array in 100 cells. DDB2-LacR tethered arrays were analyzed in a ZRF1 knockdown (shZRF1) background, after expression of either nucleolar GFP or nucleolar ZRF1ΔUBD-GFP. There was a significant difference between the DDB2-LacR tethered array in the control and mutant overexpression cells as judged by a KS test. (Middle panel) Mean nucleolar distance of the DDB2-tethered array in the corresponding cells. The mean nucleolar distance of each replicate was calculated from measurements of nucleolar distance in 100 cells. The graph shows the average of the means from 3 independent experiments ±SD. Statistical significance was determined using an unpaired t-test. (Right panel) Immunofluorescence images showing cells expressing nucleolar targeted GFP or ZRF1ΔUBD-GFP, along with DDB2-LacR. Nucleoli are marked by NPM and nuclei with DAPI. G. Knockdown of XPC abolishes repositioning of a DDB2-LacR tethered array. (Left panel) Heat map showing distribution of the nucleolar distance of the array in 100 cells. mCherry-LacR or DDB2-LacR tethered arrays were analyzed in a XPC knockdown (shXPC) background. There was no significant difference between the control and DDB2-LacR tethered array in the shXPC cells as judged by a KS test. (Right panel) Mean nucleolar distance of the control and DDB2-tethered array in shXPC cells. The mean nucleolar distance of each replicate was calculated from measurements of nucleolar distance in 100 cells. The graph shows the average of the means from 3 independent experiments ±SD. Statistical significance was determined using an unpaired t-test. (G) Hypothetical model for nucleolar tethering of damaged DNA via H2A K119-ubiquitylation.
