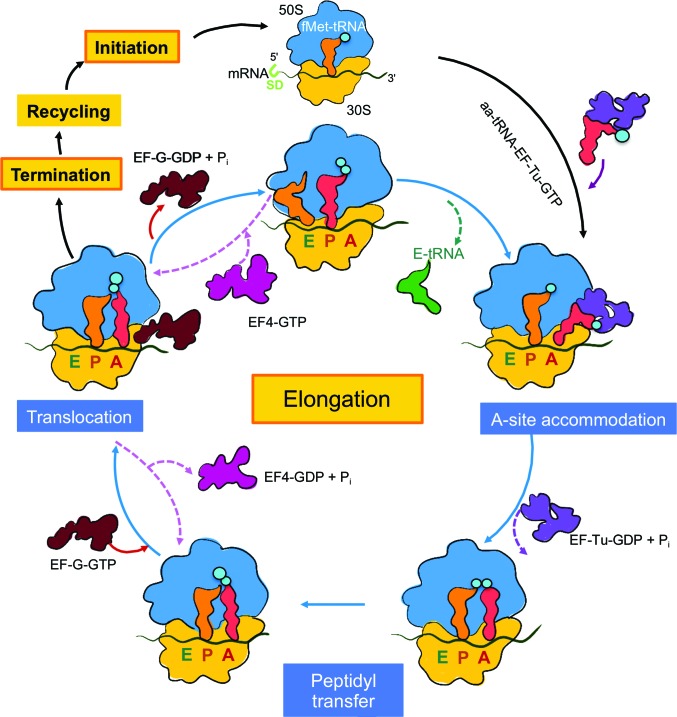
Figure 1.
Protein biosynthesis on the ribosome. The illustrated diagram shows the key protein-translation steps performed by bacterial ribosomes. During translation, the ribosome is engaged in four key steps: initiation, elongation, termination and recycling (highlighted in yellow boxes). Translation initiation starts with the 30S subunit (yellow) binding near the initiation codon on mRNA at the Shine–Dalgarno (SD) sequence (Schmeing & Ramakrishnan, 2009 ▸; light green). Upon recruitment of the formylmethionyl-tRNA (fMet-tRNA; orange) at the P-site, carrying the methionine amino acid (cyan), the 50S subunit (blue) binds to form the initiation complex. Individual steps of the elongation cycle are shown in blue boxes. An incoming aminoacyl-tRNA (aa-tRNA; red), carrying a charged amino acid (cyan circle), bound to EF-Tu-GTP (purple) binds at the A site of the ribosome (in the A-site accommodation step). Upon mRNA decoding and a correct codon–anticodon pair between the mRNA and tRNA, EF-Tu hydrolyses GTP and dislocates (shown as a purple dashed arrow), allowing peptide-bond formation between A-site and P-site tRNAs in the peptidyl-transfer step. Elongation factor EF-G (dark brown) then binds to allow tRNAs to translocate from the A to P sites and from the P to E sites (translocation step) with energy derived from GTP catalysis. The release of EF-G (GDP-bound, shows as a brown arrow) enables deacetylated tRNA to exit (E-tRNA; green). During tRNA translocation, EF4-GTP (magenta; Qin et al., 2006 ▸) can rescue stalled ribosomes by back-translocation (shown as dashed magenta arrows) to the peptidyl-transfer step to proceed with normal protein elongation.